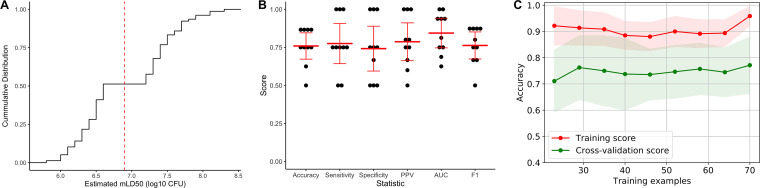
FIG 5.
Performance of the random forest algorithm in predicting P. aeruginosa virulence from accessory genomic content when intermediate virulence isolates (middle third of estimated mLD50 values) were removed. (A) Cumulative distribution function of estimated mLD50 values after removing intermediate virulence isolates. Isolates with estimated mLD50 values less than the median value in the complete training set (red dashed line) were designated high virulence, with the remainder designated low virulence. (B) Nested 10-fold cross-validation performance of the random forest model, including accuracy, sensitivity, specificity, positive predictive value (PPV), area under the receiver operating characteristic curve (AUC), and F1 score. The results for each cross-validation fold are shown in black with the mean and 95% confidence interval of each statistic indicated in red. (C) Learning curve showing change in mean training accuracy (red line) and cross-validation accuracy (green line) with increasing training set sizes. Shading indicates the 95% confidence interval. Assessments at each number of training examples were through 10-fold nested cross-validation.