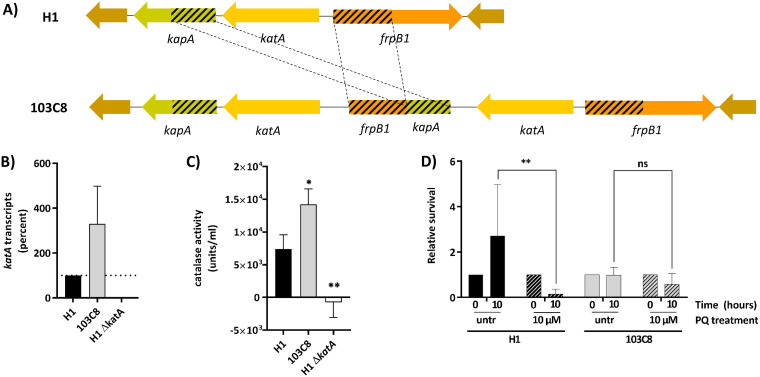
FIG 2.
In vivo rearrangement in the reisolate 103C8 due to gene fusions and duplication of the katA gene. (A) Representation of the genomic context of katA in H1 and the duplication of katA and a fusion of duplicated fragments of the genes frpB and kapA in reisolate 103C8. (B) Transcription of the catalase gene in various H. pylori clones was measured by qPCR. katA transcript is shown as % relative to the H1 reference, which was set to 100%. The reisolate 103C8 with two katA copies showed significantly higher katA transcript amounts than H1. A mutant lacking the katA gene did not show transcript, as expected. All results were normalized to the respective 16S transcript amounts, also using strain H1 as reference. (C) Catalase activity was measured in bacterial lysates of strain H1, the reisolate 103C8, and H1ΔkatA (negative control) The reisolate 103C8, with two katA copies, displayed higher catalase activity than H1. The calculation of the catalase units/ml is defined as in the Megazyme catalase assay kit instructions. Test, one-way analysis of variance (ANOVA) (P < 0.05). (D) Survival experiment using paraquat (PQ) as oxidizing agent on live bacteria. Results are shown as relative survival values, where individual data sets were normalized to their respective bacterial counts obtained at time zero hours, which were set to 1. Strains H1 and 103C8 were treated with 10 μM PQ or left untreated (untr), and the number of colonies was counted 10 h postexposure. Strain H1 was less able to resist the oxidative stress, in contrast to reisolate 103C8, which was more resistant to PQ. Test, two-way ANOVA (P < 0.05). For panel B, dotted lines refer to the duplicated and fused regions from the kapA and frpB1 genes. For panels C and D, * = P < 0.05; ** = P < 0.01; ns, not significant.