Figure 2. Relationship between genetic programs of regeneration and embryogenesis.
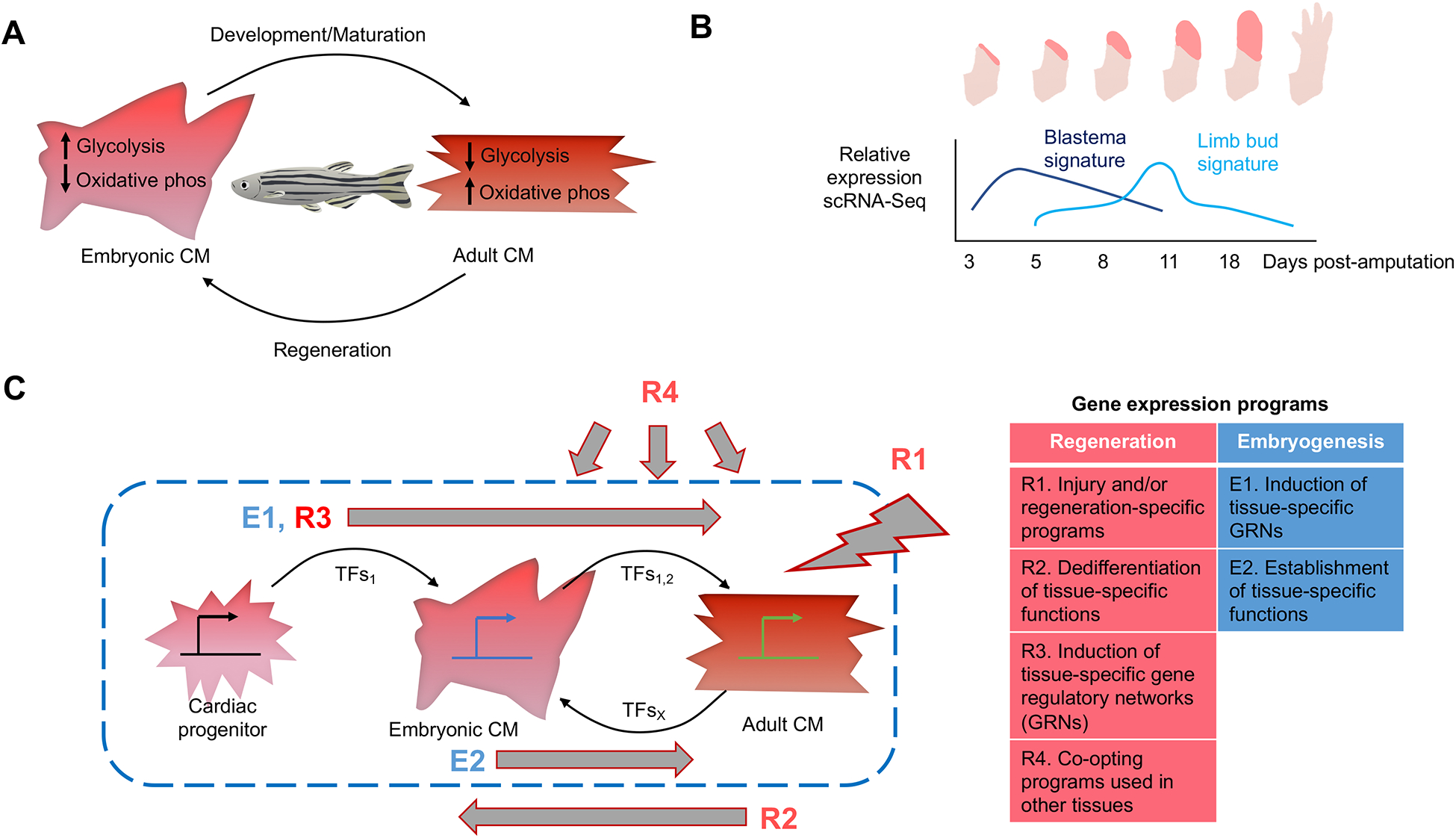
A. Heart regeneration in zebrafish is characterized by a return to certain embryonic gene expression programs17. Cardiomyocyte maturation from the embryo to the adult results in a switch from glycolytic metabolism to mitochondrial respiration (OXPHOS). During regeneration of cardiomyocytes, cells revert to a more glycolytic state through upregulating the expression of glycolytic genes B. Time series showing the biphasic gene expression response during regeneration of axolotl limbs after amputation22. Gene expression levels in sorted connective tissue cells indicate a distinct gene profile in early regenerates that is specific to blastemal generation (dark blue). A separate signature similar to that of embryonic limb buds is expressed in later regenerates (light blue). C. A model integrating gene expression programs relevant to regeneration, visualized for cardiac muscle cells. Embryonic gene programs utilize cascades of tissue-specific or tissue-preferred transcription factors, establishing tissue-specific functions. Regeneration involves defining injury signals, programs for de-differentiation of adult cell functions to approximate a less mature state, and employment of a tissue-specific gene regulatory network. Gene expression programs can also be co-opted from other tissue types or disease contexts. Transcription factors help establish and stabilize each state.
