Figure 3. Discovering DNA regulatory elements involved in regeneration.
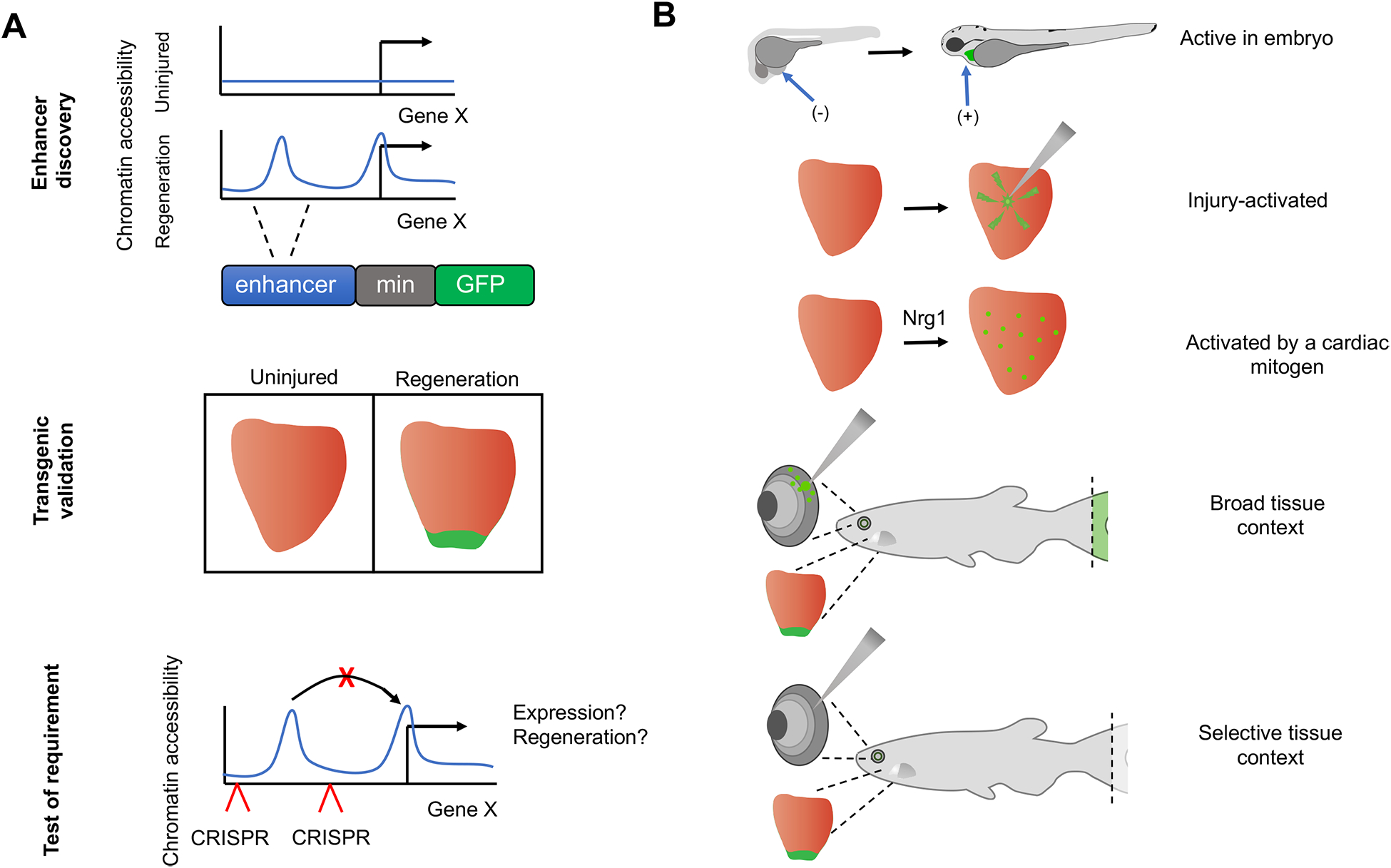
A. Tissue regeneration enhancer element (TREE) discovery typically begins with identification of a sequence region enriched for active or open chromatin marks in regenerating samples versus uninjured samples. Test DNA regions (blue) are subcloned upstream of a minimal promoter (gray) directing a reporter gene, such as GFP. In this example shown for a regenerating heart, stable transgenic lines only express GFP during regeneration. In vivo validation experiments delete the test region from the genome using CRISPR techniques and determine resulting changes in gene expression or regenerative events. B. TREEs validated in reporter experiments are activated in a context-specific manner. An enhancer might direct expression during embryonic or regenerative development. Enhancer activity can be induced by injury or by tissue creation, which can be discerned by testing expression in the presence of a mitogenic trigger such as Nrg1. Enhancers might direct expression in multiple regeneration contexts, for example regenerating fins, retinae, and heart, or instead might be tissue-specific. GFP reporter expression in represented in green.
