Figure 5. Phase transitions and injury-induced regeneration programs.
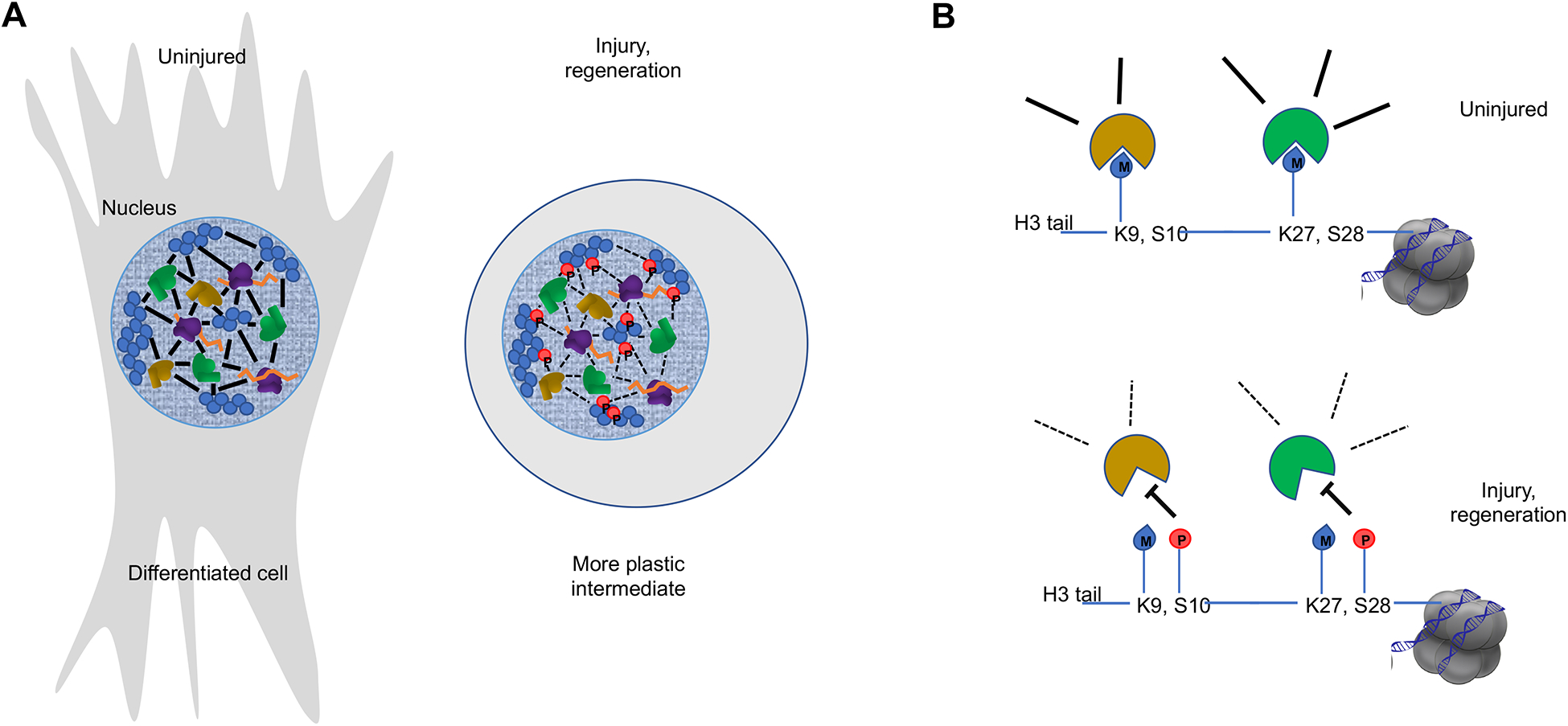
A. A model by which major phase transitions in chromatin might underlie sharp phenotypic changes during adaptive cell reprogramming. In differentiated cells, networks of transcription factors (brown), nucleosomes (blue), chromatin proteins (green), RNA (orange), and covalent modifications (not picture) coalesce into compartments of active and repressed genes. During regeneration, numerous signaling pathways are activated to help dismantle such aggregates, for example phosphorylation events (red) that inhibit binding reactions. Thus, nuclei are reset to an embryonic-like state where the genome organization is loosened, allowing for changes in cell differentiation and proliferative capacity. Chromatin binding proteins that recognize covalent modifications on histone tails, such as methylations of H3K9 and H3K27, are integral components of binding interactions that constitute a membrane-less organelle. Phosphorylation of Ser residues adjacent to the methylated Lys disrupts binding of these proteins to chromatin and could lead to phase transitions where previously structured repressed domains are released, leading to an opening of the genome.
