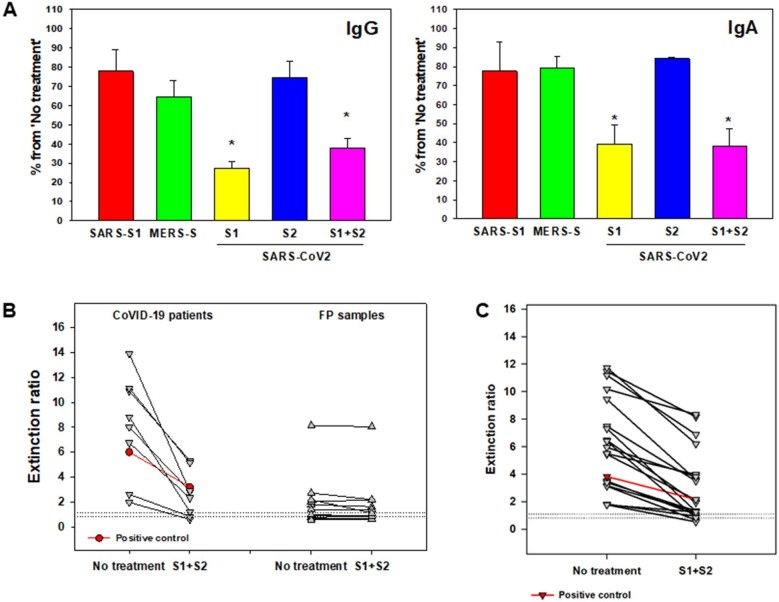
Fig. 3.
Competitive spike proteins significantly improve assay specificity. A. Nucleic acid confirmed COVID-19 patient serum specimens (n = 3) were assayed for SARS-CoV-2 IgG and IgA antibodies. Pretreatment of the sample with SARS-CoV S1 subunit protein (SARS-S1), MERS spike protein (MERS-S), SARS-CoV-2 S1 subunit protein (S1), SARS-CoV-2 S2 subunit protein (S2), and SARS-CoV-2 S1 subunit and S2 subunit (S1 + S2) was performed to assess IgG and IgA antibody reaction specificity. Results are mean ± SEM, percent of inhibition from non-treated control sample (n = 3). B. SARS-CoV-2 IgA antibodies were assessed in nucleic acid confirmed COVID-19 patients (n = 9) and non-COVID-19 samples that displayed cross-reactivity (n = 12) with and without S1 + S2. Horizonal dashed lines indicate indeterminate interpretive region. *, p < 0.01 (paired t-test). C. SARS-CoV-2 IgG antibodies were assessed in nucleic acid confirmed COVID-19 patients (n = 19) with and without S1 + S2. Horizonal dashed lines indicate indeterminate interpretive region. *, p < 0.01 (paired t-test). Positive control(s) from the kit were used as additional controls (both in B and C) to confirm spike protein specificity.