Table 5.
The best 14 compounds identified by the structure-based virtual screening approach against the SARS-CoV-2.
| DrugBank ID | 3CLpro (-8.83)a | ACE2 (-10.29)a | ACE2/spike (-7.25)a | RdRp (-11.54)a | 2D Structure | DRUG NAME |
|---|---|---|---|---|---|---|
| DB03632 | −7.29 | −9.50 | −6.97 | −10.65 | 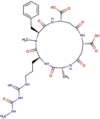 |
Argifin |
| DB01329 | −7.82 | −6.31 | −4.77 | −11.54 | 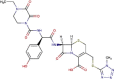 |
Cefoperazone |
| DB00430 | −7.66 | −8.84 | −4.81 | −11.07 | 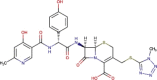 |
Cefpiramide |
| DB01415 | −6.92 | −8.88 | −4.03 | −10.07 | 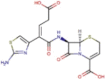 |
Ceftibuten |
| DB09050 | −7.91 | −9.15 | −6.53 | −10.74 | 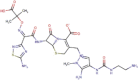 |
Ceftolozane |
| DB08995 | −6.85 | −6.86 | −6.12 | −8.07 | 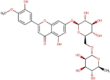 |
Diosmin |
| DB11633 | −6.85 | −5.56 | −5.30 | −7.18 | 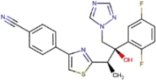 |
Isavuconazole |
| DB00722 | −6.85 | −6.68 | −5.30 | −9.88 | 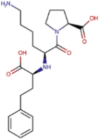 |
Lisinopril |
| DB00157 | −7.79 | −8.64 | −6.79 | −9.69 | 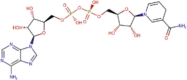 |
NADH |
| DB11871 | −7.72 | −8.57 | −6.41 | −8.23 | 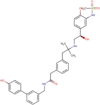 |
PF-00610355 (investigational) |
| DB121383 | −7.20 | −7.24 | −5.25 | −10.72 | 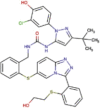 |
PF-03715455 (investigational) |
| DB14761 | −6.96 | −7.47 | −3.98 | −9.91 | 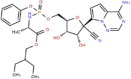 |
Remdesivir |
| DB12846 | −7.31 | −8.65 | −5.96 | −8.14 | 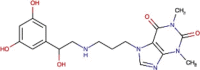 |
Reproterol |
| DB01698 | −7.81 | −6.76 | −4.54 | −9.58 | 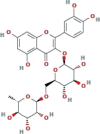 |
Rutin |
2D representation, DrugBank code, drug name and Glide score (G-Score) values of the best 14 compounds identified by the structure-based virtual screening approach against the SARS-CoV-2 main protease (3CL-PR), polymerase (RdRp) and the host ACE2 enzyme, alone and complexed to the viral spike glycoprotein. The compounds are in alphabetical order.
This value indicates the absolute best G-score value for each analyzed target and is expressed in kcal/mol.
