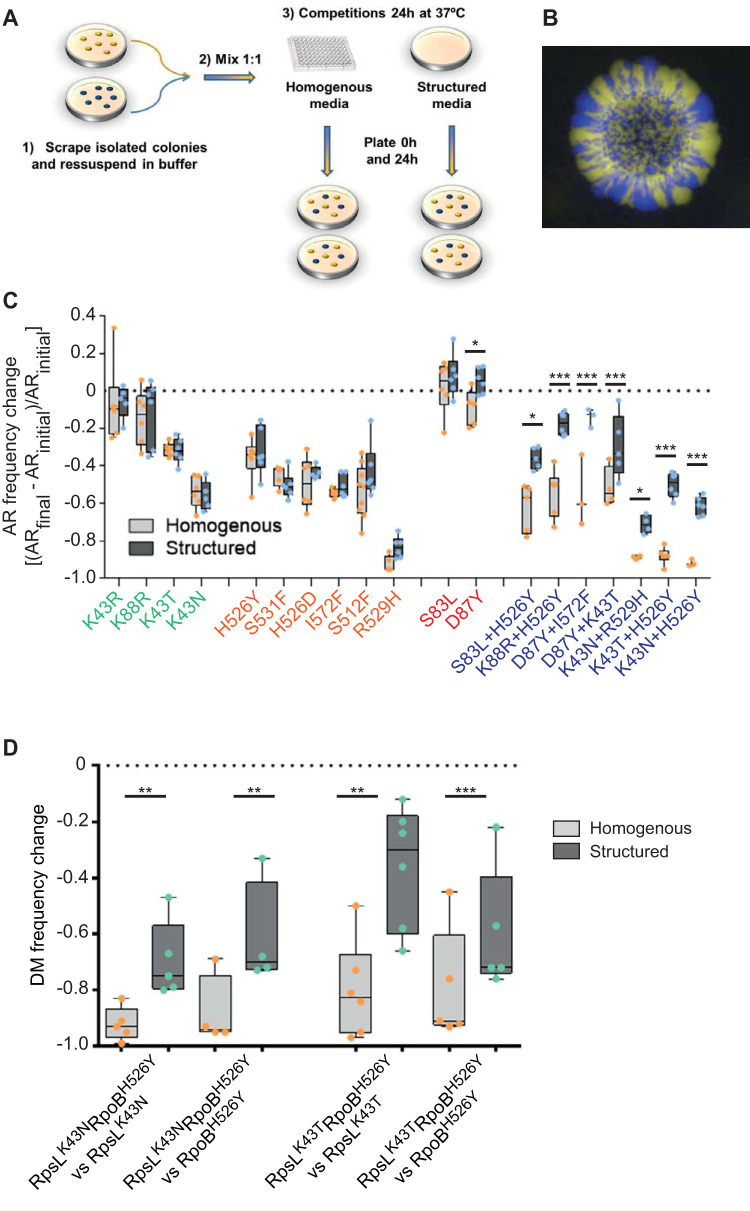
FIG 1.
Structured populations facilitate the maintenance of double antibiotic resistances. (A) Scheme of the experimental setup used to measure the frequency allele of the antibiotic resistance (AR) mutations in either liquid or 1.5% agar LB medium without antibiotics, using the susceptible strain as reference. (1) Frozen stocks were streaked to isolate single colonies from each mutant, which after overnight growth (2) were used to mix in a 1:1 ratio with the susceptible strain. (3) The same mix was used to start the competitions in homogeneous and structured populations. The initial mix (t = 0 h) and the final mix (t = 24 h) were plated with the proper dilution to count CFU/ml. For more details see Materials and Methods. (B) Example of segregation of E. coli genotypes (susceptible strain-YFP versus a susceptible strain-CFP) due to radial expansion observed in LB agar plates after 24-h competitions. (C) Frequency change of AR mutants in homogeneous populations (light gray bars) and in structured populations (dark gray bars). All the mutants competed with the susceptible background in pairwise competitions in both homogeneous and structured populations. Frequency change was calculated using the formula ([ARinitial − ARfinal]/ARinitial), where ARinitial and ARfinal stand for the frequency of the resistance mutation at time 0 h (initial mix) and after 24 h of competition, respectively. The colors in the x axis represent resistance to different antibiotics, i.e., mutations in rpsL conferring resistance to streptomycin are in green. (D) Frequency change of the double resistant mutants when competing against their single resistant counterparts in homogeneous and structured populations. The double mutant is found in a significantly higher percentage when the populations are structured.