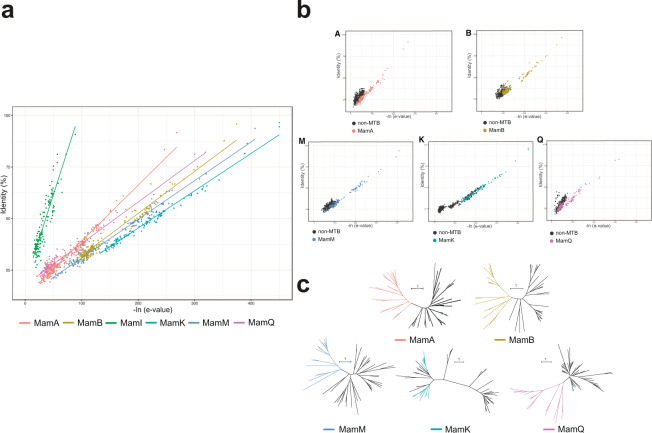
Fig. 1.
The choice of Mam protein for further searching for MGCs in open databases. (a) Correlations between –ln of e-values (x axis) and identities (y axis) among MamA, -B, -M, -K, -I, and -Q proteins sequences. (b) Correlations between identities and –ln of e-values among Mam protein sequences with their homologs. (c) Phylogenetic trees based on investigated sequences. Trees were reconstructed by the maximum-likelihood method with LG + F + I + G4 substitution model. Bootstrap values were calculated based on 1000 resamplings. Bar represents one substitution per 100 amino acid positions.