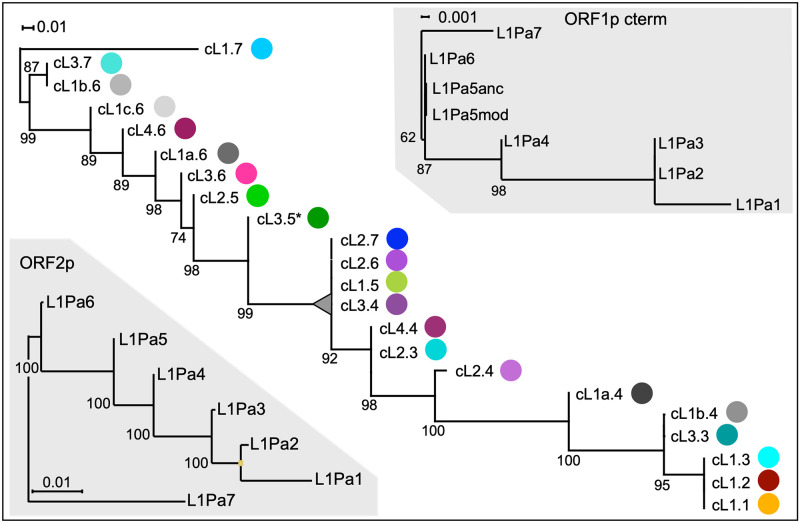
Fig 4. Coiled coil phylogeny.
Maximum likelihood trees of the coiled coil clusters and C-termini were built on their 50% consensus sequences with the amino acids encoded by CG-affected codons treated as missing data. Note the 10-fold lower scale of the branch lengths for the C-terminus tree. The colored circles at the tips of the coiled coil cluster tree correspond to the cluster colors in Fig 3, panel A-F. The numbers at each node give its frequency as % of 1000 bootstrap replicates. The ORF2 tree was generated from amino acid consensus sequences of the human version of our previously described collection of L1Pa2 –L1Pa7 human/chimpanzee orthologues [51] and the currently active human L1Pa1 family (in particular, Ta1-d 5), represented here by the L1.3 element [52]. This tree is consistent with a previously described tree built from the 3’ 2 kb of nucleic acid sequence which includes the 3’UTR but mostly ORF2 sequence (Figure 4A in [7]).