FIG 6.
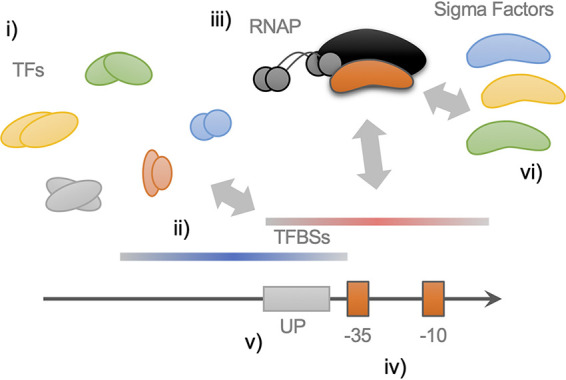
A putative model for a bacterial promoter region, including a range of experimental attributes. (i) More than 300 proteins (transcription factors [TFs]) in E. coli are predicted to bind DNA, and there is a lack of experimental characterization (1). (ii) Recently, high-throughput studies, such as genomic SELEX, are showing a large number of possible TFBS on genomes (11), which may impact on the composition of promoter sequences. These regions can have positive (blue regions) or negative (red regions) effect on promoter activity. (iii) RNAP requires a sigma factor to be recruited to the promoter sequence, and each sigma factor possesses a preference for a specific motif on DNA (1). (iv) Nucleotide composition and motifs between −10 and −35 boxes influence transcription initiation and promoter activity (46, 47). (v) The interrelation between the UP element and a subunit of RNAP were found to play a role in transcription initiation and promoter activity (44), and the UP element can switch preference of sigma factors in promoters (45). (vi) The same promoter sequence can respond to diverse sigma factors, according to experimental characterizations and in silico approaches (36, 61, 62).
