Fig. 3. Mutating the Pol α HBM impairs parental histone transfer to lagging strand in mouse ES cells.
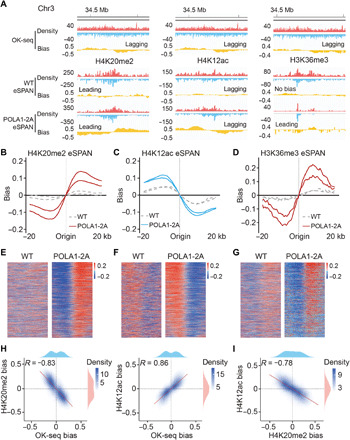
(A) Snapshots of OK-seq reads density and bias and H4K20me2, H4K12ac, and H3K36me3 eSPAN reads density and bias in WT and POLA1-2A mouse ES cells at selected origin regions on chromosome 3. (B to D) Average bias of H4K20me2 (B), H4K12ac (C), and H3K36me3 (D) eSPAN results at selected initiation zones (n = 1548) in WT and POLA1-2A mouse ES cells, each with two repeats shown. (E to G) Representative heatmap of H4K20me2 (E), H4K12ac (F), and H3K36me3 (G) eSPAN in WT and POLA1-2A mouse ES cells. (H) H4K20me2 and H4K12ac eSPAN biases in POLA1-2A mouse ES cells showed a strong anticorrelation and correlation with OK-seq bias, respectively. Spearman’s rank correlation coefficient and the density distribution were shown. (I) H4K20me2 and H4K12ac eSPAN biases in POLA1-2A mouse ES cells showed reverse correlation.
