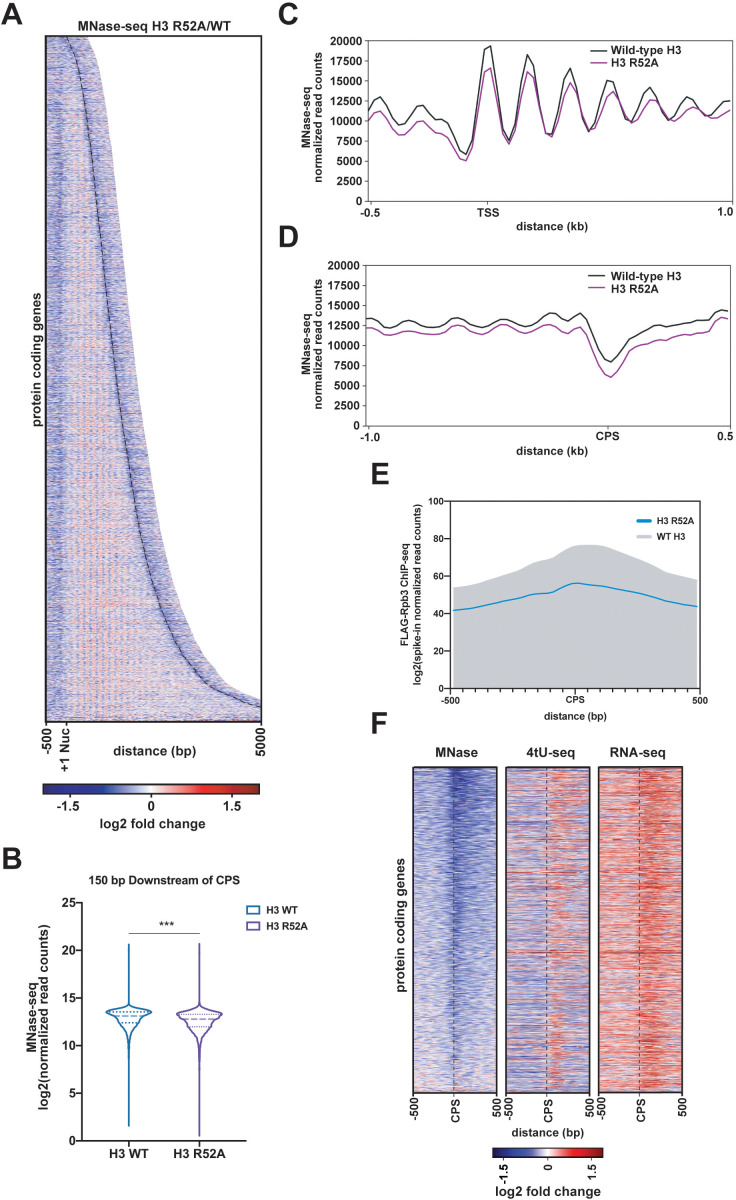
Figure 6. Mutation of the DNA entry-exit site alters nucleosomes genome-wide.
(A) Heatmap of the log2-fold change of MNase-seq read counts (spike-in normalized as described in Materials and methods) of the H3 R52A mutant relative to WT. Rows represent 6205 protein-coding genes (as in Figure 3) sorted by length and showing 500 bp upstream of the +1 nucleosome (Brogaard et al., 2012) and 500 bp downstream of the CPS (curved black dotted line; Ozsolak et al., 2010). (B) Violin plot of log2-transformed normalized read counts from MNase-seq data in the region 150 bp downstream of the CPS. The difference between WT and the H3 R52A mutant is statistically significant (p<0.0001) as determined by a Wilcoxon rank-sum test. (C) Metagene plots showing normalized MNase-seq read counts for WT (black) and the H3 R52A mutant (purple) in a region from −500 bp to +1000 bp relative to the +1 nucleosome. (D) MNase-seq metagene plots as in C, but plotted from −1000 bp to +500 bp from the CPS. (E) Metagene plots showing spike-in normalized FLAG-Rpb3 ChIP-seq read counts for wild-type (gray) and the H3 R52A mutant (blue) in a region from −500 bp to +500 bp relative to the CPS (Ozsolak et al., 2010). All heatmaps and metagene plots were generated using deepTools2 (Ramírez et al., 2014; Ramírez et al., 2016) using 25 bp bins and 6205 protein-coding genes. All MNase-seq data were produced using a 2.5 U MNase digestion. (F) Heatmaps of MNase-seq, RNA-seq and 4tU-seq data sorted (lowest to highest log2 fold change value based on the MNase-seq data) by mean row value in the MNase-seq data and centered on the CPS and extending up- and downstream by 500 bp. Rows in panel F represent 2879 protein-coding genes present on chrIV, chrVII, chrXII, chrXIV and chrXV (no evidence of aneuploidy in any genomic\transcriptomic dataset for the H3 R52A mutant).