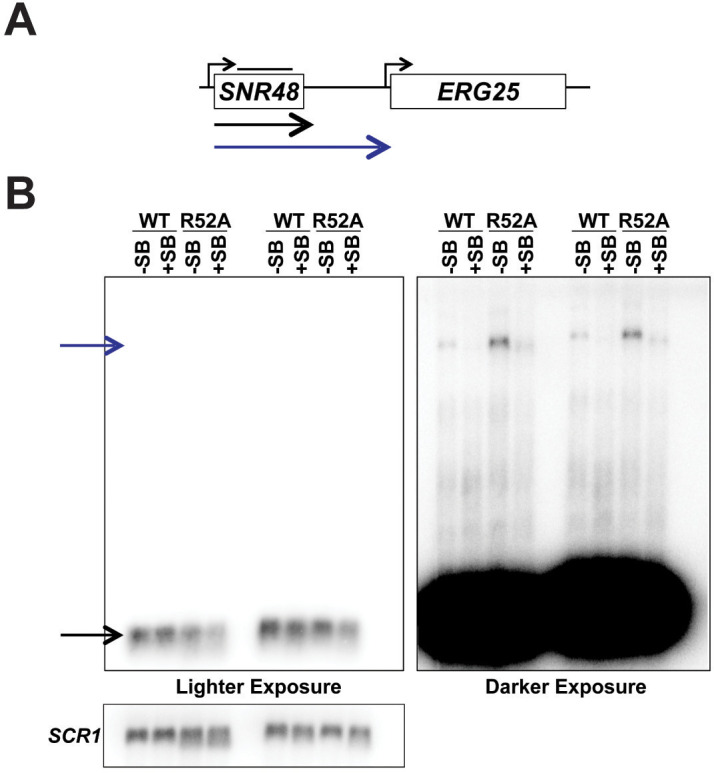
Figure 7. Integration of a superbinder DNA sequence downstream of SNR48 suppresses the termination defect of the H3 R52A mutant.
(A) MNase-seq data from a 2.5 U digestion visualized in IGV (Thorvaldsdóttir et al., 2013) (top) compared to de novo transcript annotations generated from RNA-seq data from the same strains (bottom). Arrows indicate reduced nucleosome occupancy in the H3 R52A strain compared to the wild-type, which is coincident with the 3’ end of the transcript in the wild-type strain. For scale, SNR48 is 113 bp. The site of integration of the superbinder sequence is indicated by a black bar. (B) Diagram of superbinder nucleosome with locations of the PCR primers for ChIP and the probe used in northern blot analysis to detect SNR48 readthrough transcription. (C) ChIP-qPCR using an antibody against H2A in KY3221 transformed with either wild-type or mutant histone plasmids. Error bars represent the SEM of three independent biological replicates. (D) Representative northern blot detecting the SNR48 readthrough transcript in wild-type and H3 R52A strains with or without the superbinder sequence. Northern blot analysis was performed on three independent biological replicates.
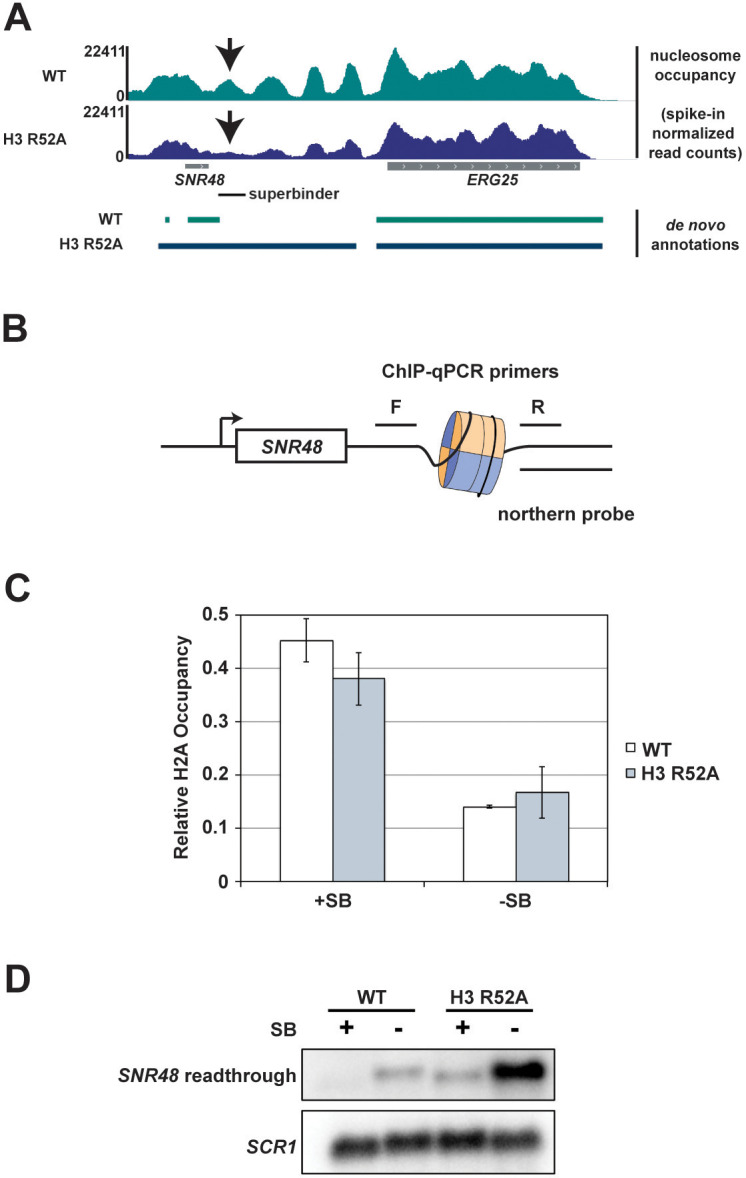
Figure 7—figure supplement 1. Detection of readthrough transcription of SNR48 by northern analysis in strains with and without the superbinder (SB) sequence.