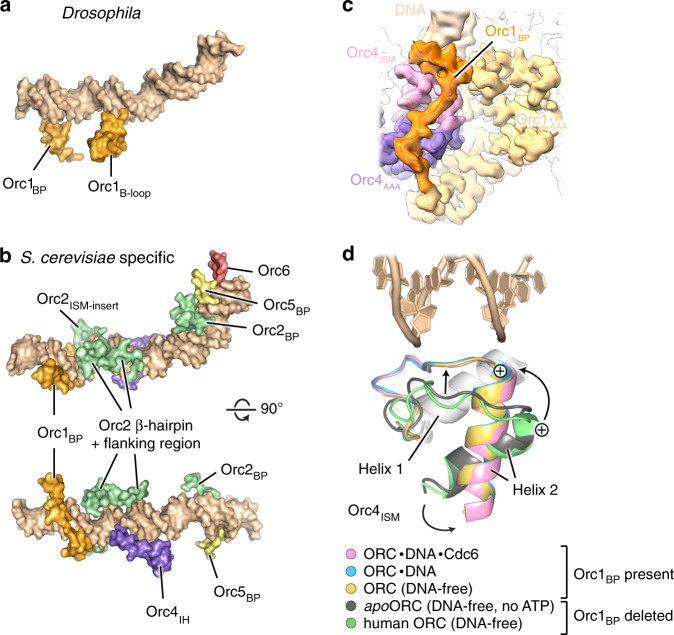
Fig. 5. Comparison of S. cerevisiae and Drosophila ORC–DNA contacts.
a The B-loop of Orc1 engages DNA in Drosophila but not in budding yeast ORC, while interactions between the Orc1-BP and DNA differ in both systems. DNA binding elements shared with S. cerevisiae ORC are not shown. b S. cerevisiae ORC uses numerous additional regions to associate with DNA in the cryo-EM structure of the DNA-bound budding yeast initiator (PDB 5zr120). A subset of these are yeast-specific insertions in ORC subunits (e.g., Orc2ISM-insert, Orc4IH), while others are flexible in DmORC·DNA and/or DmORC·DNA·Cdc6 (i.e., Orc2BP, Orc5BP, Orc6TFIIB). Only duplex-binding regions unique to the S. cerevisiae structure are shown. Note also that the basic patch of S. cerevisiae Orc1 tracks in the DNA minor groove. c In Drosophila Orc1, the basic patch packs against the ISM of Orc4 and N-terminally folds into a helix that contacts the phosphodiester backbone but does not engage the minor groove. Unsharpened EM map density is shown. d Docking of the Orc1-basic patch reorients the second Orc4-ISM helix, priming it for DNA binding. The Orc4-ISMs from DmORC structures determined in this study (all containing the Orc1-basic patch) are superposed onto the Orc4-ISMs from the apo-DmORC and human Orc1/4/5 crystal structures (PDBs 4gxc16 and 5uj717; both determined in the absence of the Orc1-basic patch). The repositioning of the positive ISM helix 2 dipole is indicated. BP, basic patch; IH, insertion helix; ISM, initiator-specific motif.