Abstract
The bifunctional human enzyme phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthetase (PAICS) catalyzes two essential steps in the de novo purine biosynthesis pathway. PAICS is overexpressed in many cancers and could be a promising target for the development of cancer therapeutics. Here, using gene knockdowns and clonogenic survival and cell viability assays, we demonstrate that PAICS is required for growth and survival of prostate cancer cells. PAICS catalyzes the carboxylation of aminoimidazole ribonucleotide (AIR) and the subsequent conversion of carboxyaminoimidazole ribonucleotide (CAIR) and l-aspartate to N-succinylcarboxamide-5-aminoimidazole ribonucleotide (SAICAR). Of note, we present the first structures of human octameric PAICS in complexes with native ligands. In particular, we report the structure of PAICS with CAIR bound in the active sites of both domains and SAICAR bound in one of the SAICAR synthetase domains. Moreover, we report the PAICS structure with SAICAR and an ATP analog occupying the SAICAR synthetase active site. These structures provide insight into substrate and product binding and the architecture of the active sites, disclosing important structural information for rational design of PAICS inhibitors as potential anticancer drugs.
Keywords: de novo purine biosynthesis, nucleotide metabolism, cancer target, rational drug design, carboxyaminoimidazole ribonucleotide (CAIR), N-succinylcarboxamide-5-aminoimidazole ribonucleotide (SAICAR), phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthetase (PAICS), prostate cancer, structural biology, purine, cancer therapy, drug design, nucleoside/nucleotide biosynthesis
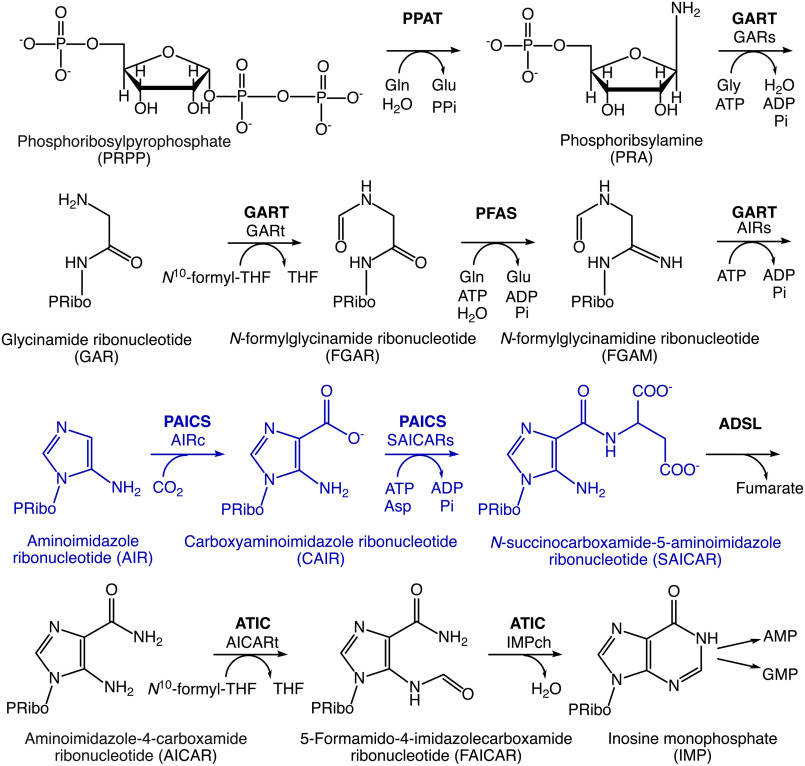
The de novo purine biosynthesis pathway, ubiquitous across the tree of life, includes a series of reactions catalyzing the conversion of phosphoribosyl pyrophosphate into inosine monophosphate, a key intermediate in nucleotide metabolism. In most eukaryotes, the de novo purine biosynthesis pathway comprises 10 enzymatic reactions (1, 2) and in humans, steps 6 and 7 are catalyzed by the bifunctional enzyme PAICS (Fig. 1). All enzymes involved in the de novo purine biosynthesis pathway are potential targets for anticancer drug design, as cancer cells are highly dependent on this pathway for proliferation, in contrast to normal cells that mainly use the nucleotide salvage pathway for purine production (3). PAICS has been reported to be overexpressed in various types of cancer such as prostate, breast, lung cancer, and melanoma, and plays a critical role in cell proliferation, invasion, and efficient tumor growth (4–11). This suggests that a good strategy to treat these cancers could be to inhibit the enzymatic activity of PAICS. Such a strategy is further supported by the finding that PAICS expression correlates with disease progression, and ability to form metastases in prostate cancer, and reduced tumor growth upon PAICS knockdown in a mouse xenograft model of prostate cancer (7). Furthermore, shRNA-mediated depletion of PAICS was shown to reduce tumor growth in mouse lung cancer and pancreas cancer models (12, 13), and to significantly hinder malignant proliferation of breast cancer cells in vitro (4) as well as in a mouse breast cancer disease model (14). Targeting PAICS has also been suggested to be a new strategy to treat neuroblastoma (15), which further supports PAICS inhibition as an interesting novel approach for rational drug design of anti-cancer therapeutics.
Figure 1.
Scheme of the de novo purine biosynthesis pathway in humans. The reactions catalyzed by PAICS are highlighted in blue. The following abbreviations are used: ADSL, adenylosuccinate lyase; AICARt, aminoimidazole-4-carboxamide ribonucleotide transformylase; ATIC, aminoimidazole-4-carboxamide ribonucleotide transformylase/inosine monophosphate cyclohydrolase; GARs, glycinamide ribonucleotide synthetase; GARt, glycinamide ribonucleotide transformylase; GART, glycinamide ribonucleotide synthetase/glycinamide ribonucleotide transformylase/aminoimidazole ribonucleotide synthetase; IMPch, inosine monophosphate cyclohydrolase; PFAS, N-formylglycinamidine ribonucleotide synthase; PPAT, phosphoribosylpyrophosphate amidotransferase; THF, tetrahydrofolate.
Steps 6 and 7 of the de novo purine biosynthesis pathway comprise the carboxylation of aminoimidazole ribonucleotide (AIR) and the subsequent ATP-dependent conversion of carboxyaminoimidazole ribonucleotide (CAIR) and l-aspartate to N-succinylcarboxamide-5-aminoimidazole ribonucleotide (SAICAR). In the majority of eukaryotic organisms these steps are catalyzed by the bifunctional enzyme PAICS (phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthetase) harboring AIR carboxylase (AIRc) and SAICAR synthetase (SAICARs) activities in a single polypeptide chain (Fig. 1), whereas prokaryotes, yeast, and plants have separate enzymes to catalyze these two reactions (16, 17). The transcription factor MYC directly regulates PAICS expression in prostate cancer (5) and a BET bromodomain inhibitor has been shown to interfere with MYC binding to the PAICS promoter and suppress PAICS gene expression (7).
Although most eukaryotes and some bacteria perform the AIRc reaction in a single step using a PurE-II–type AIRc activity, in bacteria, yeast, and plants, CAIR is synthesized in two steps: AIR is carboxylated at N5 in an ATP-dependent reaction catalyzed by PurK and the resulting N5-CAIR is then converted to CAIR by the mutase PurE-I, a homologue of PurE-II (18, 19). Structures of prokaryotic PurE-I (20–22), prokaryotic PurE-II (23), and prokaryotic (24, 25) and yeast (26–28) SAICARs (PurC) have been determined in complexes with various ligands. These structures demonstrated that PurE enzymes function as octamers whereas the prokaryotic and yeast homologues of SAICARs (PurC) are dimeric and monomeric enzymes, respectively. X-ray structures of ligand-free human (29) and insect (30) PAICS have confirmed an octameric organization also of the bifunctional enzymes, but structural information on how natural ligands bind to bifunctional PAICS is still lacking. In light of PAICS being a promising novel therapeutic target for cancer treatment, information on binding of its natural ligands is of importance for the development of potent and selective PAICS inhibitors.
We therefore set out to produce enzymatically active PAICS and to solve the structures of PAICS in complex with its natural ligands. We present here the first structures of PAICS in complex with CAIR bound in active sites of both domains and with SAICAR bound in the SAICAR synthetase domain. In addition, we report the PAICS structure with SAICAR and an ATP analog, AMP-PNP (adenosine 5′-(β,γ-imido)triphosphate), occupying the SAICAR synthetase active site. Furthermore, we confirm that PAICS depletion has a negative effect on the growth and survival of prostate cancer cells supporting the validity of PAICS as an anti-cancer target. The structural data presented here provide for the first time insight into substrate and/or product binding and the architecture of the two active sites of PAICS, thus providing important structural information for future rational design of PAICS inhibitors.
Results and discussion
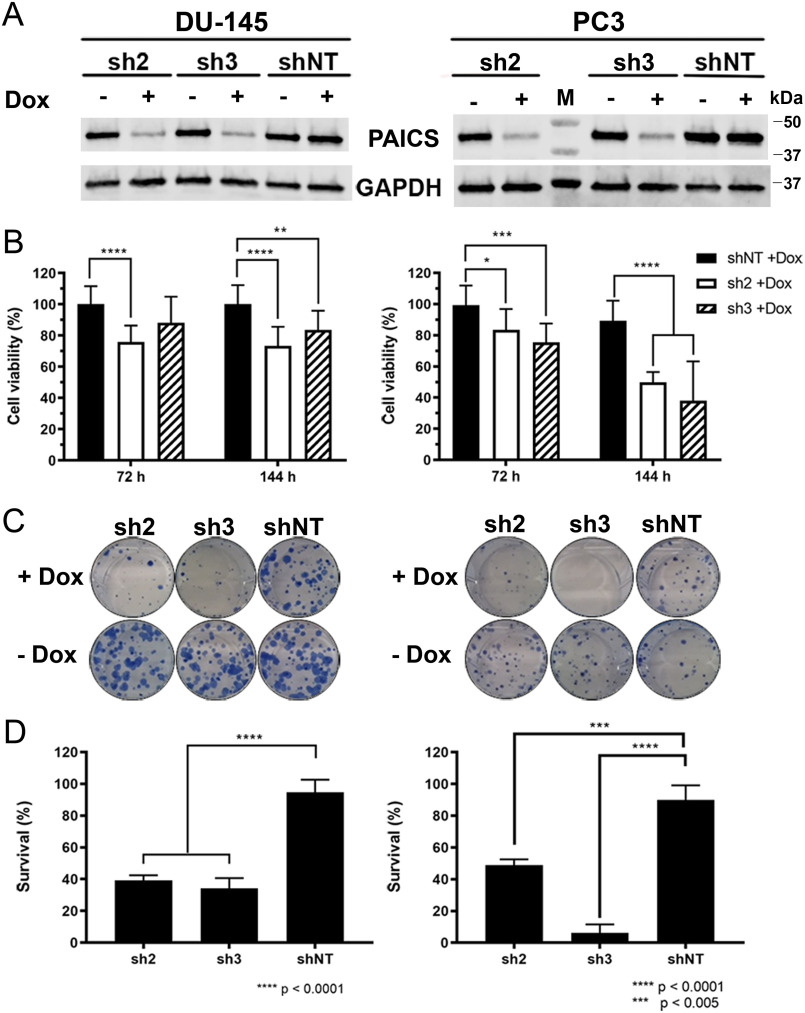
PAICS depletion reduces clonogenic survival of prostate cancer cells
To demonstrate the importance of PAICS for the growth and survival of prostate cancer cells, we have generated inducible PAICS knockdown in the androgen-independent prostate cancer cell lines PC3 and DU-145. The depletion of PAICS protein resulted in a significantly reduced ability to proliferate and form colonies in both cell lines, whereas no effect was seen in the control shNT cells (Fig. 2). These results confirm an important role of PAICS in androgen-receptor negative prostate cancer cell growth (7).
Figure 2.
The effect of PAICS knockdown on proliferation and clonogenic survival of prostate cancer cell lines. A, Western blots showing knockdown of PAICS after doxycycline induction of prostate cancer cell lines stably transfected with two different shRNAs against PAICS. B, Resazurin assay measuring short-time effects of PAICS depletion on proliferation of prostate cancer cell lines DU-145 (left panel) and PC3 (right panel). Graphs shows survival (%) normalized to shRNA NT + Dox (n = 2). C, representative plates from colony formation assay of DU-145 (left panel) and PC3 (right panel) with and without doxycycline-induced shRNA-mediated PAICS depletion. D, quantification of colony formation assay shown in C. Graphs display survival (%) of minus doxycycline control (n = 3). Statistical significance was analyzed using two-way ANOVA or ordinary one-way ANOVA in GraphPad Prism.
Recombinantly produced PAICS is enzymatically active
The PAICS protein was recombinantly expressed in Escherichia coli in fusion with His6-tagged CPD tag and was successfully purified in high yield to over 95% purity and homogeneity (Fig. S1) by a combination of affinity and size-exclusion chromatography. The activity of the purified protein was tested at different concentrations using the PAICS-catalyzed reaction converting CAIR to SAICAR, requiring l-aspartate and energy in the form of ATP, which is converted to ADP and Pi. The formed ADP was detected using an ADP-Glo Kinase Assay (Fig. S1). The amount of ADP produced clearly increased with increased concentrations of PAICS and the use of 3 nm PAICS in the activity assay shows that the enzyme is highly active and provides a robust and reproducible activity assay with a signal to background ratio of ∼20 and a Z-factor above 0.7.
Two PAICS structures with natural ligands solved
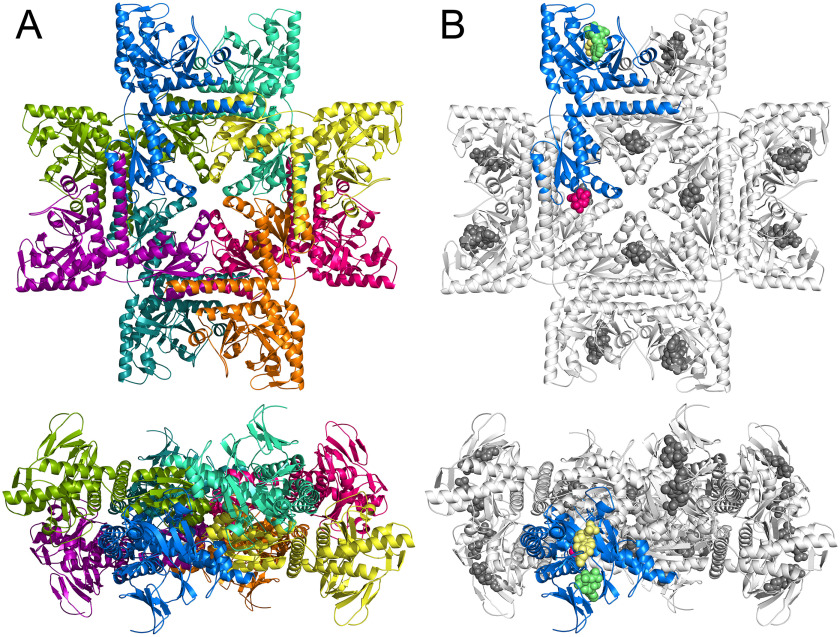
To elucidate the binding of substrates and products to PAICS, we have solved two crystal structures of PAICS in complexes with natural ligands. The data collection and refinement parameters are shown in Table 1. Because the PAICS crystals were very small thin needles, the data were collected using a microfocused high-intensity X-ray beam and the diffraction pattern exhibited anisotropy. This resulted in slightly higher Rmeas values. In one structure, determined at a resolution of 2.4 Å, both AIRc active sites of both PAICS monomers in the asymmetric unit are occupied by CAIR (the product), whereas the SAICARs active site is occupied by CAIR (the substrate) in one of the monomers and by SAICAR (the product) in the second monomer. It also contains 9 molecules of ethylene glycol and 2 imidazole molecules originating from the crystallization mother liquor. The other structure, also determined at a resolution of 2.4 Å, has empty AIRc active sites, whereas the SAICARs active sites are occupied by both SAICAR-Mg2+ (the product) and AMP-PNP-Mg2+ (an ATP substrate analog). It also contains 6 molecules of ethylene glycol. In both structures, the first six N-terminal residues could not be modeled and seem to be disordered based on the low quality of the electron density map at the N terminus. The two structures are very similar and superpose with an RMSD of 0.330 Å for the superposition of all protein atoms. Both structures have two almost identical protein chains in the asymmetric unit (RMSD of 0.242 Å and 0.182 Å for the first and second PAICS structure, respectively, for the superposition of all protein atoms). Using crystallographic symmetry operations, the biologically active PAICS octamer with a pseudo 422 symmetry (D4) was generated (Fig. 3). The PAICS octamer is composed of a tightly packed core of eight C-terminal AIRc domains surrounded by eight N-terminal SAICARs domains.
Table 1.
Crystal parameters, data collection, and refinement statistics
Statistics for the highest resolution shell are shown in parentheses.
| Ligand | CAIR, SAICAR | SAICAR, AMP-PNP, Mg2+ |
|---|---|---|
| PDB code | 6yb8 | 6yb9 |
| Data collection statistics | ||
| Space group | I222 | I222 |
| Cell parameters (Å; °) | a = 60.9, b = 153.9, c = 223.6; α = β = γ = 90 | a = 61.3, b = 154.0, c = 223.6; α = β = γ = 90 |
| Number of molecules in AU | 2 | 2 |
| Wavelength (Å) | 1 | 0.9686 |
| Resolution (Å) | 47.15-2.36 (2.51-2.36) | 47.33-2.41 (2.55-2.41) |
| Number of unique reflections | 43,523 (6,857) | 41,224 (6,531) |
| Multiplicity | 13.8 (14.3) | 10.9 (10.9) |
| Completeness (%) | 99.8 (99.2) | 99.0 (98.7) |
| Rmeas (%)a | 26.6 (119.5) | 35.3 (110.0) |
| CC(1/2) (%)b | 99.5 (77.5) | 96.6 (72.4) |
| Average I/σ(I) | 8.39 (1.81) | 5.16 (1.82) |
| Wilson B (Å2) | 37.4 | 39.7 |
| Refinement statistics | ||
| Resolution range (Å) | 47.15-2.36 (2.42-2.36) | 47.33-2.41 (2.47-2.41) |
| No. of reflections in working set | 41,346 (2,941) | 39,161 (2,788) |
| No. of reflections in test set | 2,177 (156) | 2,061 (146) |
| R value (%)c | 17.7 (32.9) | 17.7 (27.9) |
| Rfree value (%)d | 23.5 (36.6) | 22.9 (33.6) |
| RMSD bond length (Å) | 0.012 | 0.012 |
| RMSD angle (°) | 1.640 | 1.585 |
| Number of atoms in AU | 7,126 | 6,964 |
| Number of protein atoms in AU | 6,528 | 6,528 |
| Mean B value (Å2) | 39.1 | 39.2 |
| Ramachandran plot statisticse | ||
| Residues in favored regions (%) | 96.4 | 97.0 |
| Residues in allowed regions (%) | 100 | 100 |
a Rmeas defined in Ref. 31.
b Pearson's correlation coefficient determined on the data set randomly split in half.
c R value = ||Fo| − |Fc||/|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
d Rfree is equivalent to the R-value but is calculated for 5% of the reflections chosen at random and omitted from the refinement process (32).
e Determined by MolProbity (33).
Figure 3.
Overall structure of the PAICS octamer. PAICS octamer is displayed in a view along the 4-fold rotation axis (top) and rotated by 90° along the x axis (bottom). Each chain is a different color in panel A. In panel B one monomer is highlighted in blue and the ligands CAIR in the AIRc active site and SAICAR and AMP-PNP in the SAICARs active site are shown as pink, yellow, and green spheres, respectively. The CAIR originates from the PAICS-CAIR structure and the SAICAR and AMP-PNP were superposed here from the structure of the PAICS-SAICAR-Mg2+-AMP-PNP-Mg2+ complex to demonstrate the relative location of the individual binding sites in the PAICS octamer.
PAICS structure in complex with CAIR and SAICAR
The AIRc active site
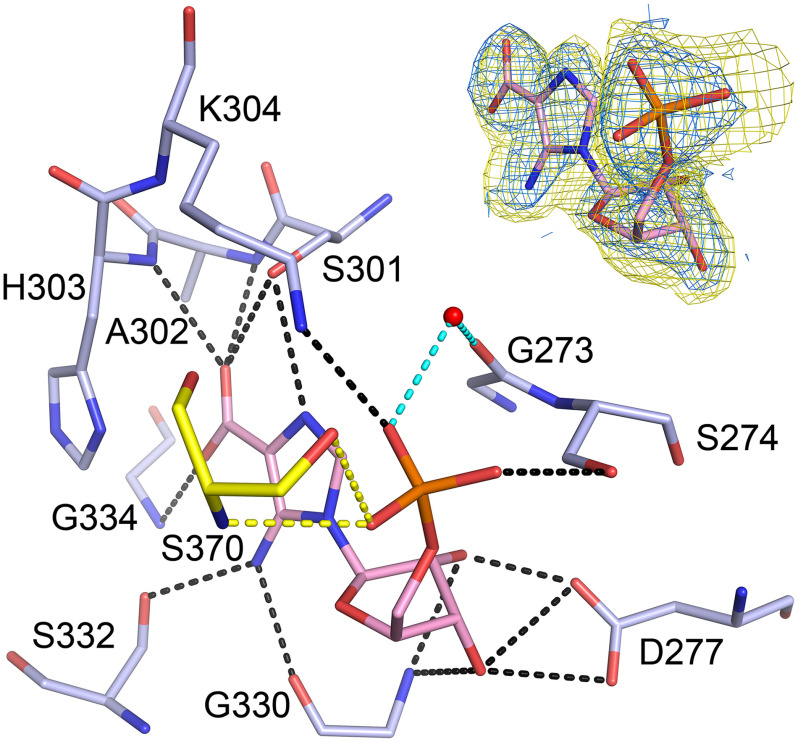
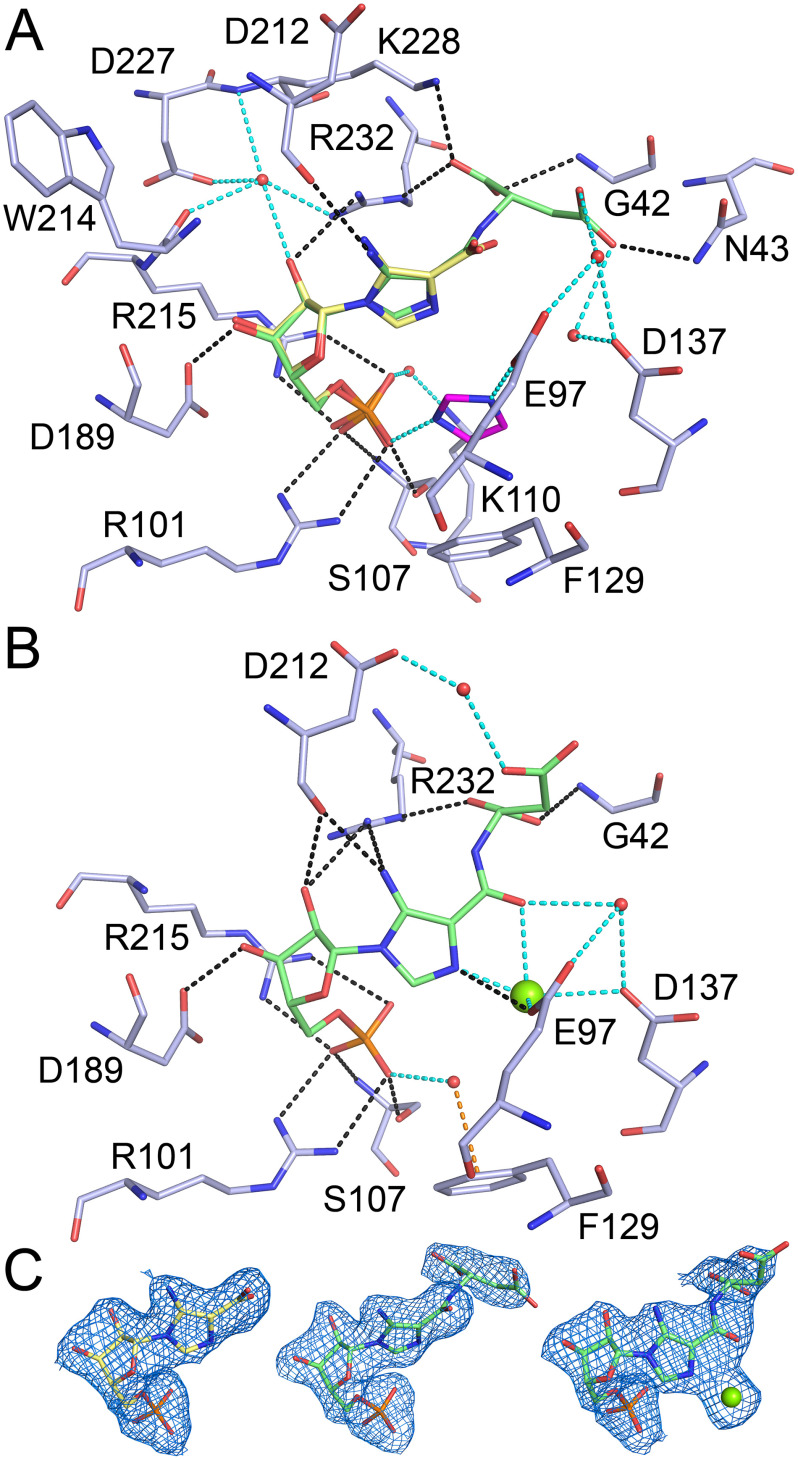
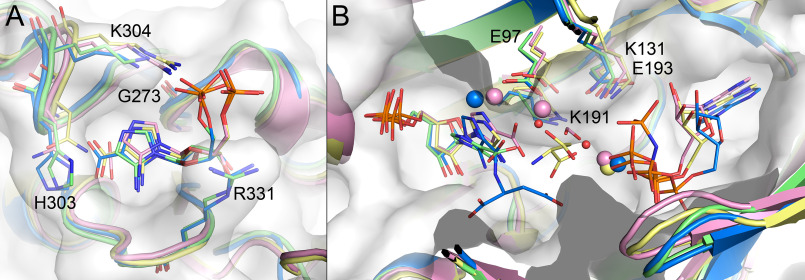
PAICS was crystallized with its final product, SAICAR, however, the SAICAR molecule appears to be unstable, as demonstrated for other intermediates in the purine de novo biosynthesis pathway like AIR and CAIR (34). SAICAR is likely to have decomposed spontaneously back to CAIR because the density in the AIRc active site corresponds to the CAIR molecule with an occupancy of 0.75. Due to the lower occupancy of the ligand and a relatively low quality of the electron density map for the ligand, we confirmed the presence of CAIR in the active site by calculating Polder omit maps for the CAIR molecule after simulated annealing refinement performed while omitting the CAIR molecules from the model (Fig. 4). The CAIR molecule, which is the product of the AIRc active site, interacts with the protein through a network of hydrogen bonds (Fig. 4). The carboxylate group of CAIR stacks with residue His-303 and residues Val-328, Ala-329, Arg-331, Asn-333, and Leu-335 are within van der Waals distance from CAIR. The phosphate moiety also interacts with residues from the neighboring PAICS monomer and forms two hydrogen bonds with residue Ser-370 and is also within van der Waals distance from residue Pro-369, which demonstrates why the octameric assembly is important for the catalytic activity of PAICS. The previously published 2.8 Å structure of human PAICS was suggested to contain a molecule of CO2 in a proposed CO2-binding site, located at the 2-fold crystallographic axis about 8 Å away from the C4 of AIR (29), which is too far for the carboxylation reaction to occur. The electron density for none of our PAICS structures suggests the presence of a CO2 molecule at this site and we propose that the actual CO2-binding site of PAICS is the one occupied in this structure by the carboxylate group of CAIR. A similar site is occupied by a CO2 molecule in the structure of the homologous enzyme PurE from Acetobacter aceti H59N inactive mutant in complex with AIR and CO2 (PDB code 5clj), suggesting that in case of human PAICS, the CO2 molecule would form main polar contacts with the main chain nitrogen atoms of residues Ala-302, Gly-334, and Leu-335.
Figure 4.
CAIR bound in the AIRc active site. The CAIR molecule is shown as pink sticks and the active site residues involved in direct (black dashed lines) or water-mediated (cyan dashed lines) hydrogen bonds are shown as pale blue sticks. Residue Ser-370 from another PAICS monomer in the octamer that participates in ligand binding through hydrogen bonds (yellow dashed lines) is shown as yellow sticks. Refined 2Fo – Fc electron density map for CAIR contoured at 1σ and mFo – DFc Polder omit map for CAIR contoured at 4 σ are shown in the upper right corner in blue and yellow, respectively.
The SAICARs active site
The SAICARs active site of the first PAICS monomer in the asymmetric unit contains a CAIR molecule, a substrate of this active site. The PAICS–CAIR interaction network of direct and water-mediated hydrogen bonds is shown in Fig. 5A. Residues Glu-97, Val-99, Thr-105, Gly-106, Met-190, Lys-191, and Ser-213 are also within van der Waals distance from the CAIR molecule. The SAICARs active site of the second PAICS monomer in the asymmetric unit contains a SAICAR molecule, which is a product of this active site (Fig. 5A). The CAIR portion of the SAICAR product forms essentially the same interaction network with the active site residues as in the case of the CAIR substrate and the N-succinyl moiety of SAICAR forms a number of additional hydrogen bond interactions (Fig. 5A), and residues Lys-17, Thr-40, Ala-41, and Asp-137 are also within the distance for van der Waals interactions. The SAICARs active sites in both PAICS monomers also contain a molecule of imidazole from the crystallization mother liquor. The imidazole molecule stacks with the imidazole ring of CAIR/SAICAR and with residue Phe-129 and forms hydrogen bonds to the phosphate of CAIR/SAICAR and to residue Glu-97. The serendipitous presence of imidazole bound in the active site might assist future design of inhibitors. The ATP-binding sites in both monomers have an ethylene glycol molecule from the crystallization mother liquor bound in the adenosine ring-binding cavity.
Figure 5.
CAIR, SAICAR, and SAICAR-Mg2+ bound in the SAICARs active site. A, the active site hydrogen bond network is shown here for SAICAR (green sticks), and the CAIR molecule (yellow sticks) from the second monomer is shown in superposition. The imidazole molecule is shown as magenta sticks. B, the active site hydrogen bond network is shown here for SAICAR-Mg2+ (green sticks and green sphere). The active site residues involved in direct (black dashed lines) or water-mediated, imidazole-mediated, and magnesium-mediated (cyan dashed lines) hydrogen bonds are shown as pale blue sticks. Residue Phe-129 that stacks with the imidazole molecule or forms a π-stacking interaction (orange dashed line) with a ligand-bound water molecule is also shown. C, refined 2Fo – Fc electron density maps for CAIR, SAICAR, and SAICAR-Mg2+ contoured at 1σ are shown left to right, respectively.
Structure of PAICS in complex with SAICAR-Mg2+ and AMP-PNP-Mg2+
The AIRc active site is not occupied by any ligand and the long binding cleft of the SAICARs active site is occupied by the product, SAICAR and AMP-PNP, which is an analog to the co-substrate ATP. Because AMP-PNP is stabilized by magnesium cations, we crystallized the enzyme in the presence of MgCl2 and the structure revealed one Mg2+ cation bound to each SAICAR and AMP-PNP molecule. These two magnesium cations together with two water molecules bridge the two ligands in the active site (Fig. 6A). Although the crystals from which the two different structures were solved were grown in very similar crystallization mother liquor, no imidazole molecules were bound in this structure.
Figure 6.
SAICARs active site with SAICAR-Mg2+ and AMP-PNP-Mg2+. A, the SAICAR-Mg2+ (left) and AMP-PNP-Mg2+ (right) molecules (green sticks and green spheres) and water molecules bridging the two ligands (red spheres) are shown in the SAICARs active pocket (blue cartoon and semi-transparent surface). B, the ATP-binding site hydrogen bond network is shown for AMP-PNP-Mg2+ (green sticks and green sphere). Active site residues involved in direct (black dashed lines) or water-mediated and magnesium-mediated (cyan dashed lines) hydrogen bonds are shown as pale blue sticks. Refined 2Fo – Fc electron density map for AMP-PNP-Mg2+ contoured at 1σ is shown in the upper left corner.
SAICAR-Mg2+ binds to the SAICARs active site with a similar interaction network as in the structure without the magnesium and AMP-PNP (Fig. 5B). The direct hydrogen-bonding pattern for the CAIR moiety is almost the same, but the ribose does not form any water-mediated interactions. Also, the imidazole ring is slightly rotated here to bind the magnesium cation, and interacts therefore directly with residue Glu-97. The phosphate moiety does not form water-mediated interactions with Lys-110, but interacts with a water molecule, which forms a π-stacking interaction with Phe-129. The amide carbonyl group of SAICAR is rotated toward the magnesium cation and interacts with a water molecule that bridges the SAICAR through another water molecule to the magnesium ion bound to AMP-PNP. Compared with the other PAICS-SAICAR structure (Fig. 5A), the N-succinyl moiety is oriented differently here, with one of the carboxyl groups pointing toward the AMP-PNP-Mg2+, and it does not interact with Asn-43 and Lys-228, but rather forms a water-mediated hydrogen bond with Asp-212 (Fig. 5B).
The AMP-PNP-Mg2+ binds next to the SAICAR molecule and forms hydrogen bond interactions mainly through the P-PNP moiety with the loop turn containing residues 17-19 and through the adenosine ring, whereas the ribose moiety does not form any major interaction with the protein (Fig. 6B). The adenosine ring is sandwiched between residues Met-93 on one side and residues Val-21, Val-31, and Leu-33 on the other side of the hydrophobic pocket and residues Tyr-14, Gly-16, Ala-88, and Gln-90 are also within van der Waals distance from AMP-PNP.
In both structures, the electron density map for the N-succinyl moiety of SAICAR is less well resolved than for the CAIR portion of SAICAR (Fig. 5C) and the B-factors for the N-succinyl atoms are higher, which might be due to the N-succinyl moiety missing in some of the unit cells and the SAICARs active site containing a mixture of CAIR and SAICAR. Alternatively, the N-succinyl moiety of SAICAR might be flexible and adopt multiple conformations. Indeed, this portion of SAICAR is oriented differently in the two structures (Fig. S2). Although we cannot rule out that the difference in the orientation is caused by the presence/absence of imidazole and magnesium, we believe that this conformational flexibility is likely to allow the final product SAICAR to leave the active site.
Intermediate tunnel system
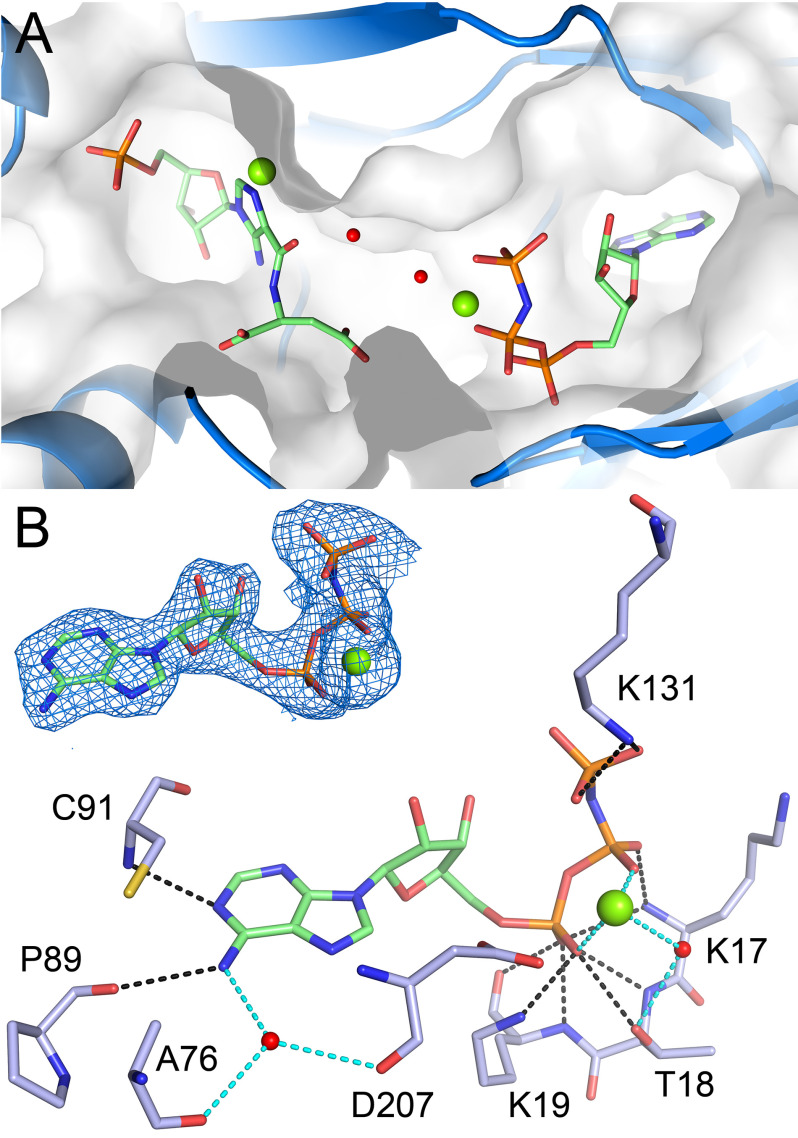
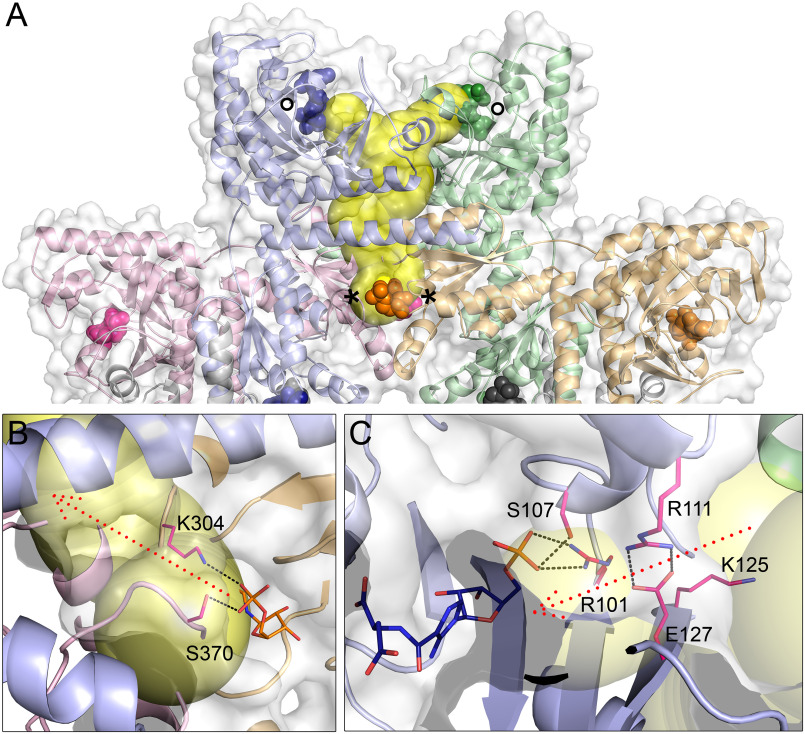
From a multifunctional enzyme efficiency perspective it would be advantageous for PAICS to channel reaction intermediates through tunnels within the protein from one active site to another without releasing the intermediate to the surrounding cytoplasm. Such transfer of reaction intermediates is common in enzymes with multiple catalytic centers (35). To investigate if the AIRc and SAICARs active sites in PAICS are interconnected via tunnels, we used the program Caver (36), and a network of tunnels that enable mutual communication of the individual active sites in the octameric assembly was identified. The shortest and most spacious communications between the two active sites are two tunnels that connect two neighboring AIRc sites with the two closest neighboring SAICARs active sites, which are located in different PAICS chains in the octamer than the AIRc sites (Fig. 7). The tunnels are inter-connected through a common “hallway” enabling the CAIR product from two AIRc sites to migrate as a substrate into two SAICARs sites through the protein interior. Four such tunnel systems exist in one PAICS octamer, each interconnecting four different PAICS monomers, which further demonstrate how the octameric assembly is important for PAICS function. This analysis shows how the ligands bound in the individual active sites are located relative to this tunnel system. Our findings about the details of this substrate channeling system are generally in agreement with the tunnel system previously described for the apo structure of human PAICS (29). The tunnel system is in most areas wide enough to accommodate the CAIR molecule without any substantial conformational changes needed. However, transient local rearrangement of side chains at the exit of the AIRc site would facilitate for CAIR to enter the hallway, and similar local changes would help CAIR enter the SAICARs site. Specifically, in the AIRc site, residues Lys-304 and Ser-370, which are involved in the binding of the phosphate of CAIR and each of them is part of a different PAICS chain, are likely to rearrange and allow CAIR to pass between them and enter the tunnel, being guided by Lys-304 (Fig. 7B). In the SAICARs site, several basic residues (Arg-101, Arg-111, Lys-125) are located along the exit of the tunnel into the active site, and are also likely to navigate CAIR into the SAICARs site by interacting with its phosphate moiety, as also suggested previously by Li et al. (29). Most importantly, CAIR is likely to interact with Arg-111. Such interaction would disrupt the bonding between Arg-111 and Glu-127 and thus allow for an efficient translocation of CAIR from the tunnel exit to the SAICARs active site, a process further assisted by Arg-101 and Ser-107 (Fig. 7C). Our structures demonstrate that some of the residues identified as important for the intermediate channeling by Li et al. (29) are directly involved in the binding of the phosphate moieties of the ligands (Lys-304, Arg-101, Arg-111; see also Figs. 4 and 5). In contrast, the inner surface of the hallway lacks positively charged residues that could potentially impede the efficient channeling of CAIR by interacting with its phosphate group. Moreover, several negatively charged residues (Glu-142, Glu-143, Asp-163) are likely to guide CAIR through the tunnel and prevent it from halting in the hallway. The residues mentioned above are conserved among bifunctional PAICS enzymes, which further supports their importance for the enzyme function.
Figure 7.
Intermediate tunnel system in PAICS. A, the tunnel connecting the AIRc (labeled with asterisks) and SAICARs (labeled with circles) active sites of 4 different PAICS monomers (differentiated by colors) in the octameric assembly is depicted in yellow, as identified by the program Caver (36). The CAIR and SAICAR molecules in the active sites are shown as spheres. Details of the AIRc site tunnel entrance and SAICARs site tunnel exit are shown in panels B and C, respectively. Key residues facilitating the entry/exit of the intermediate are shown as magenta sticks with polar interactions highlighted as black dotted lines, and the possible intermediate trajectory is shown as red dotted arrows.
Structural changes upon ligand binding
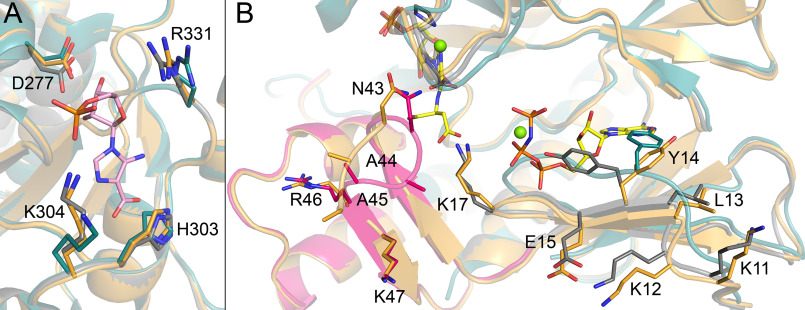
To assess ligand-induced structural changes in PAICS, we have compared our two PAICS structures to each other and also with the previously published apo structure (PDB code 2h31 (29)). The RMSD value for the superposition of the apo PAICS with both our PAICS complexes with native ligands is 0.9 Å for 386 aligned residues. When comparing the AIRc site in our two PAICS structures, the local conformation of the active site is the same with the exception of slight changes in the positions of some side chains. Because SAICAR decomposes to CAIR, we cannot exclude that the AIRc active site in the structure of PAICS with SAICAR-Mg2+ and AMP-PNP-Mg2+ contains a small amount of CAIR too, despite that the electron density map does not suggest so. Therefore, we also performed a structural comparison with the apo PAICS structure, which clearly indicates that the AIRc active site is pre-formed and binding of CAIR into the AIRc active site indeed does not trigger any major conformational changes and only small side chain rearrangements occur. For example, side chains of residues Lys-304 and Arg-331 move inwards in the active site, the ring of His-303 flips outwards from the active site to accommodate the carboxylate moiety of CAIR, and the side chain of residue Asp-277 is oriented slightly closer to CAIR to form hydrogen bonds with the hydroxyl groups of the ribose moiety; the carboxyl carbon of Asp-277 is shifted by 0.8 Å as compared with our structure with the empty AIRc site and by 1.2 Å relative to the previously published apo structure (Fig. 8A). Residues Pro-369 and Ser-370 from the neighboring PAICS monomer are not affected by CAIR binding.
Figure 8.
Ligand-induced structural changes in PAICS active sites. Cartoon representations of PAICS in complex with CAIR, SAICAR-Mg2+, and AMP-PNP-Mg2+, and without any ligand (PDB code 2h31 (29)) are overlaid in gray, orange, and cyan, respectively. Side chains of residues located in the positions structurally affected by ligand binding are shown in sticks with residue numbers indicated. A, AIRc site is shown with CAIR in pink stick representation. B, SAICARs site is shown with CAIR and imidazole molecules in gray sticks and with SAICAR-Mg2+ and AMP-PNP-Mg2+ in yellow sticks and green spheres. The portion of the protein that is unstructured in the apo structure and folds in the presence of CAIR is highlighted in pink in the otherwise gray cartoon of the PAICS-CAIR complex.
The SAICARs site exhibits more profound structural changes. Upon the binding of AMP-PNP the N-terminal β-sheet (residues 13-15) is moved outwards from the AMP-PNP–binding site and the side chain of Tyr-14 flips away from the binding site, not to clash with the ribose-phosphate moiety (see Fig. 8B). In case of the apo PAICS structure, this region forms a loop (residues 12-19) that is located inside the ATP-binding site with Tyr-14 penetrating into the binding site for the adenosine ring. This suggests that the ATP-binding site is already pre-formed upon the binding of the other substrate, CAIR. A part of the structure in the CAIR-binding region is largely unstructured in the apo PAICS structure (residues 36-51 and 221-238 are missing in the model) (29) and folds upon the binding of CAIR. This portion of the enzyme is folded in the previously reported structure of insect bi-functional octameric PAICS (PDB code 4ja0 (30), RMSD 0.9 Å for 401 residues), because two sulfate molecules from the crystallization conditions are bound in the SAICARs active site, mimicking the phosphate of CAIR and the β-phosphate of ATP (30). The loop comprising residues 41-46 is oriented differently in the CAIR-bound and SAICAR-bound SAICARs sites, irrespective of the presence or absence of AMP-PNP in the ATP-binding subsite. The loop might change its conformation upon the conversion of CAIR to SAICAR, but because we were not able to grow diffracting crystals and therefore not able to solve the structure with both CAIR and aspartate bound in the SAICARs site, we cannot draw any conclusions on the relevance of these observed structural changes.
Comparison with structures of PAICS homologues
AIR carboxylase domain
The only structure of a class II PurE enzyme in complex with any ligand is Treponema denticola PurE II, a prokaryotic AIR carboxylase that has been crystallized in complex with AIR (PDB code 3rgg, RMSD 0.9 Å for 154 residues) (23). Prokaryotic class I PurE enzymes, which catalyze the conversion of N5-CAIR to CAIR through a PurE-AIR-CO2 intermediate (22), are also homologous to the AIRc domain of PAICS and structures of their complexes with various ligands are available (20–22). Fig. 9A shows the superposition of the AIRc domain of PAICS with homologous PurE enzymes in complexes with relevant ligands. The superposition demonstrates that PAICS binds CAIR in the AIRc domain in the same manner as T. denticola PurE II binds the substrate AIR. Superposition of PAICS with the structure of A. aceti PurE I (catalytically inactive mutant H59N) in complex with the reaction intermediates, AIR and CO2 (PDB code 5clj, RMSD 2.1 Å for 153 residues), and with E. coli PurE I (catalytically inactive mutant H45N) in complex with CAIR (PDB code 2nsl, RMSD 2.2 Å for 157 residues) (21) indicates that the aminoimidazole ring of CAIR in PAICS overlays well with that of CAIR in E. coli PurE I and is located closer to the inside (bottom) of the cavity than the aminoimidazole ring of AIR in both class I and class II PurE. The C2 carbon of the aminoimidazole ring of CAIR in the human PAICS is located further away from the rear wall of the active site (Gly-273) than in the class I purE enzymes, similarly to the class II purE from T. denticola. This structural information supports the previously suggested position of the portal through which the CO2 molecule enters the CO2/carboxylate-binding site located deepest in the AIRc pocket, being restricted by Gly-273 and C2 of AIR (23). The phosphate moiety of the CAIR molecule in the E. coli PurE I is oriented slightly more outwards of the active site than in the case of the PurE II-type enzymes PAICS and T. denticola PurE II. The carboxylate moieties of the CAIR molecules coincide with each other and also with the CO2 molecule. Differentially conserved, class-specific residues (22, 23) involved in CAIR binding are also shown in Fig. 9A (Lys-304 in PAICS and PurE II versus R in PurE I and Arg-341 in PAICS and PurE II versus G in PurE I).
Figure 9.
Ligand binding in PAICS compared with homologues. A, AIRc site of PAICS in complex with CAIR (blue) is shown in superposition with prokaryotic PurE II from T. denticola in complex with AIR (green, PDB code 3rgg (23)), with PurE I from A. aceti in complex with AIR and CO2 (pink, H59N catalytically inactive mutant, PDB code 5clj), and with PurE I from E. coli in complex with CAIR (yellow, H45N catalytically inactive mutant, PDB code 2nsl (21)). Differentially conserved, class-specific residues involved in CAIR binding, His-303, and Gly-273 (and equivalent residues) are shown as sticks. B, SAICARs site of PAICS in complex with SAICAR-Mg2+ and AMP-PNP-Mg2+ (blue) and in complex with CAIR (green) is shown in superposition with SAICARs (PurC) from E. coli in complex with CAIR, ADP, formate, and magnesium cations (pink, PDB code 2gqs (24)) and with SAICARs (PurC) from S. pneumoniae in complex with AIR, ADP, aspartate, and magnesium cations (yellow, PDB code 4fe2 (25)). Side chains of suggested catalytic residues are shown as sticks. Semitransparent surface representation of PAICS is also shown in both panels.
In conclusion, the comparison of the structures of the AIRc domain of human bifunctional PAICS and its homologues, prokaryotic PurE enzymes from class I and class II, demonstrates that ligand binding to these enzymes is very similar. The difference lies mainly in the position of the phosphate moiety, caused by the differentially conserved phosphate-binding residue, Lys-304 in human PAICS and other class II PurE enzymes, versus an arginine in the case of PurE I enzymes. Despite this slight variance, the molecular details of ligand binding to these enzymes remain highly conserved throughout the tree of life.
SAICAR synthetase domain
Structures of both prokaryotic (24, 25) and eukaryotic (26–28) homologues of the SAICARs domain of PAICS in complexes with various ligands are available. CAIR or AIR together with ADP and magnesium cations can be found in the structures of SAICARs (PurC) from E. coli (PDB code 2gqs (24), RMSD 1.5 Å for 232 residues) and Streptococcus pneumoniae (PDB code 4fe2 (25), RMSD 1.5 Å for 234 residues), respectively. Fig. 9B shows the superposition of the PAICS SAICARs site with these homologous complexes. In our PAICS structures the imidazole ring of CAIR is positioned at a different angle than that of SAICAR, which might be caused by the lack of magnesium cations in the PAICS-CAIR structure. Otherwise the CAIR and SAICAR align relatively well with the CAIR and AIR in the homologous structures. The N-succinyl moiety of SAICAR does not bind to the substrate-binding site for aspartate, but extends outwards from the binding cleft, allowing for the final product SAICAR to exit the active site. The aspartate-binding site is occupied by two water molecules in our structure of PAICS with SAICAR-Mg2+ and AMP-PNP-Mg2+, and by a formate molecule and an additional magnesium cation in the E. coli structure. AMP-PNP-Mg2+ adopts a bent conformation and aligns well with the ADP-Mg2+ in the homologous structures (Fig. 9B). In the two available structures of ATP-bound prokaryotic homologues (Pyrococcus horikoshii, PDB code 4o7w (37), RMSD 1.6 Å for 226 residues and Methanocaldococcus jannaschii, PDB code 2z02, RMSD 1.7 Å for 233 residues), ATP is also present in a bent conformation. The bent ATP conformation is consistent with the proposed catalytic mechanism that involves transfer of the γ-phosphate moiety from ATP to Glu-193, then to Glu-97 and finally to the carboxylate of CAIR, to activate it for the reaction with the aspartate positioned near the vicinity of the activated carboxylate group of CAIR; the phosphate group charge is compensated for by Lys-131, Lys-191, and magnesium cations (25). These key residues are highly conserved and adopt the same conformation in our human PAICS structures as in the homologues shown in Fig. 9B.
Complexes of SAICARs from Saccharomyces cerevisiae with various ligands have been reported, which include SAICAR (PDB code 2cnv), aspartate (PDB code 2cnu), and ATP (PDB code 1obd), with RMSD values of 2.7 Å for 233 aligned residues (26–28). In these structures, however, the aspartate and the N-succinyl moiety of SAICAR bind to a different site than in the human PAICS and prokaryotic PurC, further away from the proposed active site residues. Moreover, the ATP is bound in the ATP-binding pocket in an extended conformation, in contrast to AMP-PNP in the human PAICS structure and its prokaryotic homologues.
In summary, the binding poses of the ligands in the SAICARs domain of human bifunctional PAICS are very similar to the ligand poses in the prokaryotic homologues. The ligand binding in the eukaryotic homologue from yeast is different, but we chose not to draw any conclusion from this, as the S. cerevisiae ligand-bound structures do not seem to be compatible with the proposed catalytic mechanism of these enzymes. In contrast, the comparison with the prokaryotic PurC enzymes indicates that our structures present catalytically relevant complexes.
We show that depletion of PAICS greatly impedes the growth of androgen-independent prostate cancer cells as demonstrated by proliferation and colony formation assays, further supporting PAICS as a valid target for the treatment of prostate cancer. We successfully expressed and purified catalytically active PAICS and here present the first structures of PAICS in complex with its natural ligands. Each structure provides detailed information on the molecular architecture of the active sites and also on ligand binding by PAICS. Both binding pockets, especially the CAIR- and ATP-binding sites of the SAICARs active site, display various types of interactions with the bound ligands, in particular hydrophobic, stacking, polar, and charge–charge interactions. These offer a chemical space that can be exploited for the design of high-affinity specific inhibitors. The structural data and the enzyme activity assay reported here will be useful in future structure-based rational design of potential anti-cancer drugs targeting PAICS.
Experimental procedures
PAICS knockdown and clonogenic survival assay
Generation of stable cell lines
Prostate cancer cell lines PC3 and DU-145 were obtained from ATCC and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, at 37 °C in 5% CO2 humidified atmosphere. Inducible knockdown cell lines were prepared by lentiviral delivery of anti-PAICS shRNA in an inducible pRSITEP vector system following a previously published procedure (38). In brief, custom forward and reverse oligos (Sigma Aldrich, Table S1) were annealed and ligated into pRSITEP vector (Addgene), digested by FastDigest BshTI and EcoRI, before transforming into One Shot Stbl3 Chemically Competent Cells (Invitrogen No. C737303) according to the manufacturer's protocol. Validation of correct inserts was performed by sequencing. Stable lentiviruses carrying anti-PAICS shRNA were produced by transfecting HEK293T cells with a mixture of shRNA vector and packaging plasmids (12260 and 12259; Addgene). Virions were harvested after 36 and 60 h post-transfection and used to transduce PC3 and DU-145 cells, followed by selection with 1 μg/ml of puromycin for 7 days.
Clonogenic survival assay
Cells were seeded at densities of 200-500 cells/well in 6-well plates. After 24 h media on the cells was changed to fresh media with or without 1 μg/ml of doxycycline. Media was changed after 6 days and cells were fixed after a total of 12 days with Carnoy's fixative (methanol:acetic acid, 3:1, v/v) and stained with 0.4% crystal violet. Cells were counted and cell survival determined by referring to the number of cells seeded and normalized to the survival of respective cells without doxycycline treatment.
Cell viability assay
Cells were seeded at 500 cells/well into clear 96-well cell culture plates and left to settle for 18 h at 37 °C, 5% CO2. The following day fresh medium with 1 μm doxycycline (inducing knockdown) or without (100% viability control) was added to the cells. Cells were grown at 37 °C for 72 h before medium was replaced by fresh medium ± 1 μm doxycycline. After another 72 h of cell growth, resazurin, pre-diluted in PBS to 0.06 mg/ml, was added to the cells to a final concentration of 0.01 mg/ml. Cells were incubated for 6 h at 37 °C before reading the fluorescence at excitation/emission 540/580 nm using a Hidex Sense Microplate Reader. The fluorescence signal was corrected for the signal detected in the absence of cells.
Protein expression, purification, and activity assay
An expression construct of human PAICS in pET22b-CPDSalI (Addgene Plasmid No. 38251) was produced. Expression from this plasmid results in a PAICS construct C terminally fused to the Vibrio cholerae MARTX toxin cysteine protease domain (CPD) C terminally tagged with a His6 tag. The human PAICS protein was expressed in the E. coli strain BL21 (DE3) T1R pRARE2 at 18 °C overnight after induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Bacteria were harvested by centrifugation and lysed using sonication in lysis buffer (100 mm HEPES, 500 mm NaCl, 10% glycerol, 10 mm imidazole, 0.5 mm TCEP, pH 8.0). The lysate was cleared by centrifugation and the filtered supernatant loaded on a HisTrap HP column (GE Healthcare). After washing the column with HisTrap wash buffer (20 mm HEPES, 500 mm NaCl, 10% glycerol, 50 mm imidazole, 0.5 mm TCEP, pH 7.5), the HisTrap column with bound protein was incubated in HisTrap wash buffer supplemented with 500 μm inositol hexakisphosphate (No. P8810, Sigma Aldrich) overnight at 4 °C. PAICS protein was eluted with gel filtration buffer (25 mm HEPES, 100 mm NaCl, 0.5 mm TCEP, 0.5% glycerol, pH 7.5) supplemented with 20 mm imidazole. The eluted PAICS protein was concentrated and loaded onto a HiLoad 16/60 Superdex 200 (GE Healthcare) gel filtration column. Collected fractions were analyzed using SDS-PAGE and fractions containing PAICS were pooled, aliquoted, concentrated to 38 mg/ml, and stored at −80 °C in storage buffer (25 mm HEPES, 100 mm NaCl, 0.5 mm TCEP, 5% glycerol, pH 7.5).
PAICS activity was assayed by detecting ADP formed in the reaction step producing SAICAR from CAIR and l-aspartate through hydrolysis of ATP. PAICS (3, 30, or 300 nm) was incubated for 40 min at 22 °C in white 384-well assay plates (Corning No. 3574) with 180 μm l-aspartate (Sigma Aldrich, No. A9256), 10 μm CAIR (a kind gift from Stephen Firestine, Wayne State University), and 30 μm ultrapure ATP (included in the Kinase Assay from Promega No. V9101) in PAICS assay buffer (50 mm Tris-HCl, pH 8.0, 10 mm DTT, 0.01% BSA, 0.01% Tween 20, 20 mm MgCl2). The reaction was started by the addition of ATP and l-aspartate to the reaction well. Formed ADP was detected using ADP-Glo Kinase Assay (Promega No. V9101) according to the manufacturer's recommendations by measuring luminescence. Controls in which PAICS or l-Asp/ATP were replaced with buffer were included. An ADP standard curve was included on the plate to convert luminescence to the concentration of ADP.
Crystallization and diffraction data collection
The crystals were obtained by sitting-drop vapor diffusion technique at 21 °C in conditions from the Morpheus protein crystallization screen (39). The protein storage buffer was 25 mm HEPES, 100 mm NaCl, 0.5 mm TCEP, 5% glycerol, pH 7.5.
To obtain crystals of the PAICS-SAICAR/CAIR complex, the protein at the concentration of 16 mg/ml was supplemented with 5 mm SAICAR (No. S688795, Toronto Research Chemicals), incubated 20 min on ice, and clarified by centrifugation (14,000 × g, 10 min, 4 °C). 90 nl of the protein sample were mixed with 90 nl of the reservoir solution containing 0.1 m MES/imidazole, pH 6.5, 10% (w/v) PEG 8000, 20% (v/v) ethylene glycol, 0.02 m sodium l-glutamate, 0.02 m dl-alanine, 0.02 m glycine, 0.02 m dl-lysine HCl, and 0.02 m dl-serine. Small needle-shaped crystals (approximately 20 × 10 × 150 μm) grew overnight and were harvested after 1 week. A complete diffraction data set at 2.36 Å was collected at 100 K at the beamline X06SA at the Swiss Light Source and processed using the XDS software (40).
To obtain the crystals of PAICS in complex with SAICAR, AMP-PNP, and Mg2+, 6 mg/ml of protein was supplemented with 5 mm SAICAR (No. S688795, Toronto Research Chemicals), 5 mm AMP-PNP (No. A2647, Sigma Aldrich), and 10 mm MgCl2, incubated 20 min on ice, and clarified by centrifugation (14,000 × g, 10 min, 4 °C). 450 nl of the protein sample were mixed with 450 nl of the reservoir solution containing 0.1 m MES/imidazole, pH 6.5, 10% (w/v) PEG 8000, 20% (v/v) ethylene glycol, 0.02 m sodium formate, 0.02 m ammonium acetate, 0.02 m trisodium citrate, 0.02 m sodium potassium l-tartrate, and 0.02 m sodium oxamate. Crystals of the same size and morphology as for the PAICS-CAIR complex grew overnight and were harvested after 4 days. A complete diffraction data set at 2.41 Å was collected at 100 K at the beamline i24 of the Diamond Light Source and processed using the XDS software (40).
The structures were solved by molecular replacement using the program Molrep (41) and the apo structure of human PAICS (PDB code 2h31) as a search model. Structure refinement was performed using the program Refmac 5.8.0232 (42) from the CCP4 package (43) in combination with manual adjustments in Coot software (44). MolProbity server (33) was used for evaluation of the final model quality. The presence of CAIR in the AIRc active site in the final refined model was validated by simulated annealing refinement with phenix.refine (45), whereas omitting the CAIR molecules, followed by the calculation of Polder omit maps for the CAIR molecules (solvent exclusion radius 3.0) (46). The data collection and refinement statistics are listed in Table 1. All the figures representing structures were created using PyMOL (47) and the presence of tunnels in the structure were analyzed using the Caver plugin for PyMOL (36). The RMSD values for PAICS homologues were obtained from the Dali server (48).
Data availability
Atomic coordinates and structure factors were deposited in the PDB under the accession codes 6YB8 and 6YB9.
Supplementary Material
Acknowledgments
We thank Oskar Aurelius, Hugo Lebrette, and Jonathan Davies for suggestions about crystallographic data processing and refinement and Juliane John and Riccardo Diamanti for help with X-ray data collection. We thank the Diamond Light Source for beamtime (proposal mx15806), and the staff of beamline I24 for assistance with crystal testing and data collection. The crystallographic data collection on the X06SA beamline at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland, and the assistance of the local staff is gratefully acknowledged. We thank Dr. Steven Firestine at the Wayne State University for kindly supplying the CAIR substrate, M. Altun for providing the pRSITEP vector system, and the Protein Science Facility at the Karolinska Institute for protein production.
This article contains supporting information.
Author contributions—J. S., J. U., M. G., and A-S. J. formal analysis; J. S. validation; J. S., J. U., M. G., and A-S. J. investigation; J. S., J. U., and A-S. J. visualization; J. S., J. U., and A-S. J. methodology; J. S., J. U., and A-S. J. writing-original draft; J. S., J. U., P. M., A-S. J., and P. S. writing-review and editing; E. H., A-S. J., and P. S. conceptualization; E. H., T. H., A-S. J., and P. S. supervision; E. H., T. H., A-S. J., and P. S. project administration; T. H. and P. S. funding acquisition; P. S. resources.
Funding and additional information—This work was supported by Swedish Research Council Grants 2015-00162 (to T. H.) and 2018-03406 (to P. S.), Swedish Cancer Society Grants CAN 2018/0658 (to T. H.) and CAN 2017/716 (to P. S.), Torsten and Ragnar Söderberg Foundation and European Research Council Grant TAROX-695376 (to T. H.). The work of J.S. was supported from ERDF/ESF, OP RDE, project “IOCB Mobility” (No. CZ.02.2.69/0.0/0.0/16_027/0008477) granted to the Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences. The work of P. S. was further supported by the Crafoord foundation.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- PAICS
- phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthetase
- AIR
- aminoimidazole ribonucleotide
- AIRc
- AIR carboxylase
- AMP-PNP
- adenosine 5′-(β,γ-imido)triphosphate
- CAIR
- carboxyaminoimidazole ribonucleotide
- SAICAR
- N-succinylcarboxamide-5-aminoimidazole ribonucleotide
- SAICARs
- SAICAR synthetase
- CPD
- cysteine protease domain
- shRNA
- short hairpin RNA
- RMSD
- root mean square deviation
- Dox
- doxycycline
- ANOVA
- analysis of variance
- TCEP
- tris(2-carboxyethyl)phosphine.
References
- 1. Hartman S. C., and Buchanan J. M. (1959) Biosynthesis of the purines: XXVI. the identification of the formyl donors of the transformylation reactions. J. Biol. Chem. 234, 1812–1816 [PubMed] [Google Scholar]
- 2. Lukens L. N., and Buchanan J. M. (1959) Biosynthesis of the purines: XXIV. the enzymatic synthesis of 5-amino-1-ribosyl-4-imidazolecarboxylic acid 5'-phosphate from 5-amino-1-ribosylimidazole 5'-phosphate and carbon dioxide. J. Biol. Chem. 234, 1799–1805 [PubMed] [Google Scholar]
- 3. Yin J., Ren W., Huang X., Deng J., Li T., and Yin Y. (2018) Potential mechanisms connecting purine metabolism and cancer therapy. Front. Immunol. 9, 1697 10.3389/fimmu.2018.01697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meng M., Chen Y., Jia J., Li L., and Yang S. (2018) Knockdown of PAICS inhibits malignant proliferation of human breast cancer cell lines. Biol Res 51, 24 10.1186/s40659-018-0172-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barfeld S. J., Fazli L., Persson M., Marjavaara L., Urbanucci A., Kaukoniemi K. M., Rennie P. S., Ceder Y., Chabes A., Visakorpi T., and Mills I. G. (2015) Myc-dependent purine biosynthesis affects nucleolar stress and therapy response in prostate cancer. Oncotarget 6, 12587–12602 10.18632/oncotarget.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chakravarthi B., Rodriguez Pena M. D. C., Agarwal S., Chandrashekar D. S., Hodigere Balasubramanya S. A., Jabboure F. J., Matoso A., Bivalacqua T. J., Rezaei K., Chaux A., Grizzle W. E., Sonpavde G., Gordetsky J., Netto G. J., and Varambally S. (2018) A role for de novo purine metabolic enzyme PAICS in bladder cancer progression. Neoplasia 20, 894–904 10.1016/j.neo.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chakravarthi B. V., Goswami M. T., Pathi S. S., Dodson M., Chandrashekar D. S., Agarwal S., Nepal S., Hodigere Balasubramanya S. A., Siddiqui J., Lonigro R. J., Chinnaiyan A. M., Kunju L. P., Palanisamy N., and Varambally S. (2017) Expression and role of PAICS, a de novo purine biosynthetic gene in prostate cancer. Prostate 77, 10–21 10.1002/pros.23243 [DOI] [PubMed] [Google Scholar]
- 8. Cifola I., Pietrelli A., Consolandi C., Severgnini M., Mangano E., Russo V., De Bellis G., and Battaglia C. (2013) Comprehensive genomic characterization of cutaneous malignant melanoma cell lines derived from metastatic lesions by whole-exome sequencing and SNP array profiling. PLoS ONE 8, e63597 10.1371/journal.pone.0063597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eißmann M., Schwamb B., Melzer I. M., Moser J., Siele D., Köhl U., Rieker R. J., Wachter D. L., Agaimy A., Herpel E., Baumgarten P., Mittelbronn M., Rakel S., Kögel D., Böhm S., et al. (2013) A functional yeast survival screen of tumor-derived cDNA libraries designed to identify anti-apoptotic mammalian oncogenes. PLoS ONE 8, e64873 10.1371/journal.pone.0064873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Serão N. V., Delfino K. R., Southey B. R., Beever J. E., and Rodriguez-Zas S. L. (2011) Cell cycle and aging, morphogenesis, and response to stimuli genes are individualized biomarkers of glioblastoma progression and survival. BMC Med. Genomics 4, 49 10.1186/1755-8794-4-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun W., Zhang K., Zhang X., Lei W., Xiao T., Ma J., Guo S., Shao S., Zhang H., Liu Y., Yuan J., Hu Z., Ma Y., Feng X., Hu S., et al. (2004) Identification of differentially expressed genes in human lung squamous cell carcinoma using suppression subtractive hybridization. Cancer Lett. 212, 83–93 10.1016/j.canlet.2004.03.023 [DOI] [PubMed] [Google Scholar]
- 12. Goswami M. T., Chen G., Chakravarthi B. V., Pathi S. S., Anand S. K., Carskadon S. L., Giordano T. J., Chinnaiyan A. M., Thomas D. G., Palanisamy N., Beer D. G., and Varambally S. (2015) Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer. Oncotarget 6, 23445–23461 10.18632/oncotarget.4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Agarwal S., Chakravarthi B. V., Kim H. G., Gupta N., Hale K., Balasubramanya S. A. H., Oliver P. G., Thomas D. G., Eltoum I. E. A., Buchsbaum D. J., Manne U., and Varambally S. (2020) PAICS, a de novo purine biosynthetic enzyme, is overexpressed in pancreatic cancer and is involved in its progression. Transl. Oncol. 13, 100776 10.1016/j.tranon.2020.100776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gallenne T., Ross K. N., Visser N. L., S., Desmet C. J., Wittner B. S., Wessels L. F. A., Ramaswamy S., and Peeper D. S. (2017) Systematic functional perturbations uncover a prognostic genetic network driving human breast cancer. Oncotarget 8, 20572–20587 10.18632/oncotarget.16244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheung C. H. Y., Hsu C. L., Tsuei C. Y., Kuo T. T., Huang C. T., Hsu W. M., Chung Y. H., Wu H. Y., Hsu C. C., Huang H. C., and Juan H. F. (2019) Combinatorial targeting of MTHFD2 and PAICS in purine synthesis as a novel therapeutic strategy. Cell Death Dis 10, 786 10.1038/s41419-019-2033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Firestine S. M., and Davisson V. J. (1994) Carboxylases in de novo purine biosynthesis: characterization of the Gallus gallus bifunctional enzyme. Biochemistry 33, 11917–11926 10.1021/bi00205a030 [DOI] [PubMed] [Google Scholar]
- 17. Firestine S. M., Poon S. W., Mueller E. J., Stubbe J., and Davisson V. J. (1994) Reactions catalyzed by 5-aminoimidazole ribonucleotide carboxylases from Escherichia coli and Gallus gallus: a case for divergent catalytic mechanisms. Biochemistry 33, 11927–11934 10.1021/bi00205a031 [DOI] [PubMed] [Google Scholar]
- 18. Meyer E., Kappock T. J., Osuji C., and Stubbe J. (1999) Evidence for the direct transfer of the carboxylate of N5-carboxyaminoimidazole ribonucleotide (N5-CAIR) to generate 4-carboxy-5-aminoimidazole ribonucleotide catalyzed by Escherichia coli PurE, an N5-CAIR mutase. Biochemistry 38, 3012–3018 10.1021/bi9827159 [DOI] [PubMed] [Google Scholar]
- 19. Mueller E. J., Meyer E., Rudolph J., Davisson V. J., and Stubbe J. (1994) N5-carboxyaminoimidazole ribonucleotide: evidence for a new intermediate and two new enzymatic activities in the de novo purine biosynthetic pathway of Escherichia coli. Biochemistry 33, 2269–2278 10.1021/bi00174a038 [DOI] [PubMed] [Google Scholar]
- 20. Constantine C. Z., Starks C. M., Mill C. P., Ransome A. E., Karpowicz S. J., Francois J. A., Goodman R. A., and Kappock T. J. (2006) Biochemical and structural studies of N5-carboxyaminoimidazole ribonucleotide mutase from the acidophilic bacterium Acetobacter aceti. Biochemistry 45, 8193–8208 10.1021/bi060465n [DOI] [PubMed] [Google Scholar]
- 21. Hoskins A. A., Morar M., Kappock T. J., Mathews I. I., Zaugg J. B., Barder T. E., Peng P., Okamoto A., Ealick S. E., and Stubbe J. (2007) N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis. Biochemistry 46, 2842–2855 10.1021/bi602436g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mathews I. I., Kappock T. J., Stubbe J., and Ealick S. E. (1999) Crystal structure of Escherichia coli PurE, an unusual mutase in the purine biosynthetic pathway. Structure 7, 1395–1406 10.1016/S0969-2126(00)80029-5 [DOI] [PubMed] [Google Scholar]
- 23. Tranchimand S., Starks C. M., Mathews I. I., Hockings S. C., and Kappock T. J. (2011) Treponema denticola PurE Is a bacterial AIR carboxylase. Biochemistry 50, 4623–4637 10.1021/bi102033a [DOI] [PubMed] [Google Scholar]
- 24. Ginder N. D., Binkowski D. J., Fromm H. J., and Honzatko R. B. (2006) Nucleotide complexes of Escherichia coli phosphoribosylaminoimidazole succinocarboxamide synthetase. J. Biol. Chem. 281, 20680–20688 10.1074/jbc.M602109200 [DOI] [PubMed] [Google Scholar]
- 25. Wolf N. M., Abad-Zapatero C., Johnson M. E., and Fung L. W. (2014) Structures of SAICAR synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg2+, AIR and Asp. Acta Crystallogr. D Biol. Crystallogr. 70, 841–850 10.1107/S139900471303366X [DOI] [PubMed] [Google Scholar]
- 26. Antonyuk S. V., Grebenko A. I., Levdikov V. M., Urusova D. V., Melik-Adamyan V. R., Lamzin V. S., and Wilson K. S. (2001) X-ray diffraction study of the complexes of SAICAR synthase with adenosine triphosphate. Crystallogr. Rep. 46, 620–625 10.1134/1.1387127 [DOI] [Google Scholar]
- 27. Urusova D. V., Antonyuk S. V., Grebenko A. I., Lamzin V. S., and Melik-Adamyan V. R. (2003) X-ray diffraction study of the complex of the enzyme SAICAR synthase with substrate analogues. Crystallogr. Rep. 48, 763–767 10.1134/1.1612597 [DOI] [Google Scholar]
- 28. Urusova D. V., Levdikov V. M., Antonyuk S. V., Grebenko A. I., Lamzin V. S., and Melik-Adamyan V. R. (2006) X-ray diffraction study of the complex of the enzyme SAICAR synthase with the reaction product. Crystallogr. Rep. 51, 824–827 10.1134/S1063774506050129 [DOI] [Google Scholar]
- 29. Li S. X., Tong Y. P., Xie X. C., Wang Q. H., Zhou H. N., Han Y., Zhang Z. Y., Gao W., Li S. G., Zhang X. C., and Bi R. C. (2007) Octameric structure of the human bifunctional enzyme PAICS in purine biosynthesis. J. Mol. Biol. 366, 1603–1614 10.1016/j.jmb.2006.12.027 [DOI] [PubMed] [Google Scholar]
- 30. Taschner M., Basquin J., Benda C., and Lorentzen E. (2013) Crystal structure of the invertebrate bifunctional purine biosynthesis enzyme PAICS at 2.8 A resolution. Proteins 81, 1473–1478 10.1002/prot.24296 [DOI] [PubMed] [Google Scholar]
- 31. Diederichs K., and Karplus P. A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275 10.1038/nsb0497-269 [DOI] [PubMed] [Google Scholar]
- 32. Brunger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 10.1038/355472a0 [DOI] [PubMed] [Google Scholar]
- 33. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Groziak M. P., Bhat B., and Leonard N. J. (1988) Nonenzymatic synthesis of 5-aminoimidazole ribonucleoside and recognition of its facile rearrangement. Proc. Natl. Acad. Sci. U.S.A. 85, 7174–7176 10.1073/pnas.85.19.7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang X., Holden H. M., and Raushel F. M. (2001) Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu. Rev. Biochem. 70, 149–180 10.1146/annurev.biochem.70.1.149 [DOI] [PubMed] [Google Scholar]
- 36. Pavelka A., Sebestova E., Kozlikova B., Brezovsky J., Sochor J., and Damborsky J. (2016) CAVER: algorithms for analyzing dynamics of tunnels in macromolecules. IEEE/ACM Trans. Comput. Biol. Bioinform. 13, 505–517 10.1109/TCBB.2015.2459680 [DOI] [PubMed] [Google Scholar]
- 37. Manjunath K., Jeyakanthan J., and Sekar K. (2015) Catalytic pathway, substrate binding and stability in SAICAR synthetase: a structure and molecular dynamics study. J. Struct. Biol. 191, 22–31 10.1016/j.jsb.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 38. Eshtad S., Mavajian Z., Rudd S. G., Visnes T., Boström J., Altun M., and Helleday T. (2016) hMYH and hMTH1 cooperate for survival in mismatch repair defective T-cell acute lymphoblastic leukemia. Oncogenesis 5, e275 10.1038/oncsis.2016.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gorrec F. (2009) The MORPHEUS protein crystallization screen. J. Appl. Crystallogr. 42, 1035–1042 10.1107/S0021889809042022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vagin A., and Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 10.1107/S0021889897006766 [DOI] [Google Scholar]
- 42. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 43. Collaborative Computational Project N. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 44. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 45. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liebschner D., Afonine P. V., Moriarty N. W., Poon B. K., Sobolev O. V., Terwilliger T. C., and Adams P. D. (2017) Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 73, 148–157 10.1107/S2059798316018210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schrodinger L. L. C. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrodinger, LLC, New York [Google Scholar]
- 48. Holm L., and Sander C. (1995) Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 10.1016/S0968-0004(00)89105-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors were deposited in the PDB under the accession codes 6YB8 and 6YB9.