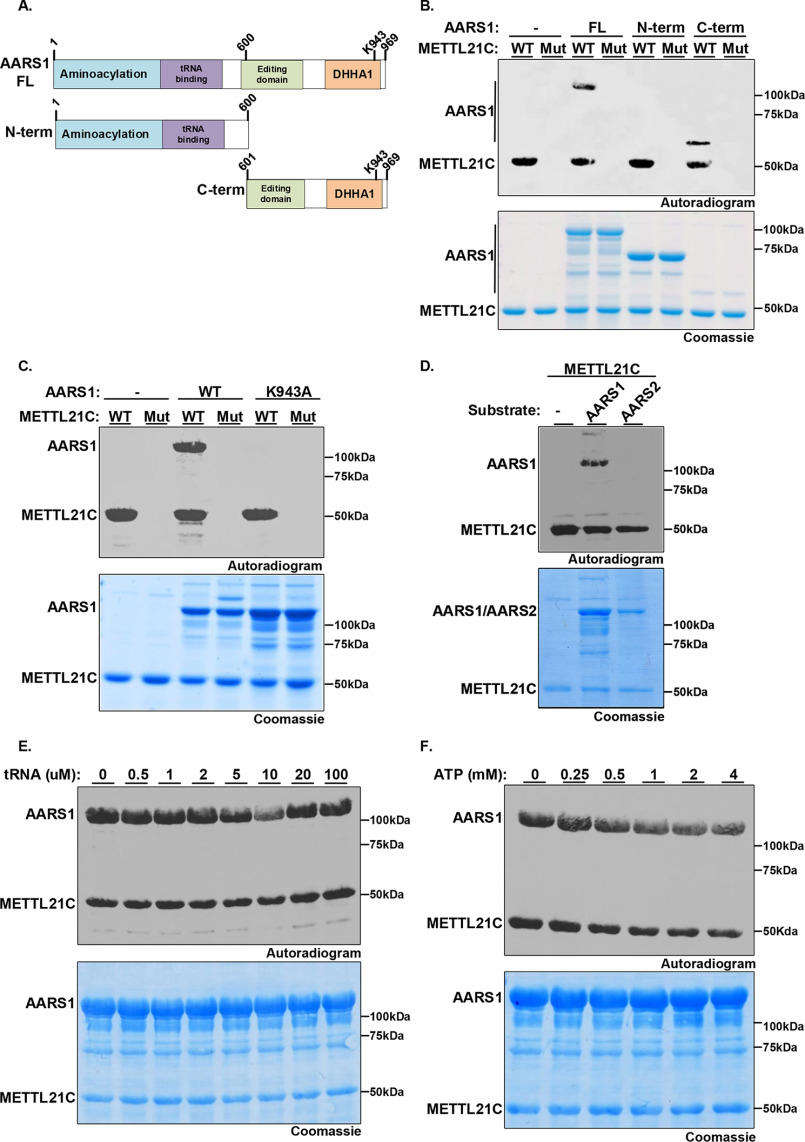
Figure 2.
METTL21C specifically methylates AARS1 at Lys-943. A, schematic diagram of AARS1 protein including functional domains and the two derivatives used to map site of methylation: N-term, the N-terminal region (1-600 amino acids) containing the aminoacylation domain and the tRNA-binding domain; C-term, the C-terminal region (601-969 amino acids) containing the editing domain and oligomerization DHHA1 domain. AARS1 full-length is named AARS1-FL. B, the C-terminal region of AARS1 is necessary and sufficient to serve as substrate for METTL21C. In vitro methylation assays as shown in Fig. 1B with WT or Mut METTL21C as indicated on AARS1 FL, N-term, or C-term as indicated. Top panel, autoradiogram. Bottom panel, Coomassie stain of proteins in the reactions. C, METTL21C specifically methylates AARS1 at Lys-943. In vitro methylation assays as in B with the indicated enzyme and substrate. D, METTL21C methylates AARS1 but not AARS2. In vitro methylation assays as in B with METTL21C on substrates AARS1 or the mitochondrial proteins AARS2. E and F, tRNA and ATP do not impact on METTL21C methylation activity on AARS1. In vitro methylation assays as in B using METTL21C and AARS1 as substrate in the presence of tRNA (E) or ATP (F) at the indicated concentrations.