Abstract
The Nem1-Spo7 complex in the yeast Saccharomyces cerevisiae is a protein phosphatase that catalyzes the dephosphory-lation of Pah1 phosphatidate phosphatase, required for its translocation to the nuclear/endoplasmic reticulum membrane. The Nem1–Spo7/Pah1 phosphatase cascade plays a major role in triacylglycerol synthesis and in the regulation of phospholipid synthesis. In this work, we examined Spo7, a regulatory subunit required for Nem1 catalytic function, to identify residues that govern formation of the Nem1-Spo7 complex. By deletion analysis of Spo7, we identified a hydrophobic Leu-Leu-Ile (LLI) sequence comprising residues 54–56 as being required for the protein to complement the temperature-sensitive phenotype of an spo7Δ mutant strain. Mutational analysis of the LLI sequence with alanine and arginine substitutions showed that its overall hydrophobicity is crucial for the formation of the Nem1-Spo7 complex as well as for the Nem1 catalytic function on its substrate, Pah1, in vivo. Consistent with the role of the Nem1–Spo7 complex in activating the function of Pah1, we found that the mutational effects of the Spo7 LLI sequence were on the Nem1–Spo7/Pah1 axis that controls lipid synthesis and related cellular processes (e.g. triacylglycerol/phospholipid synthesis, lipid droplet formation, nuclear/endoplasmic reticulum membrane morphology, vacuole fusion, and growth on glycerol medium). These findings advance the understanding of Nem1-Spo7 complex formation and its role in the phosphatase cascade that regulates the function of Pah1 phosphatidate phosphatase.
Keywords: phospholipid, phospholipid metabolism, triglyceride, protein phosphatase, phosphatidate, phosphatase, membrane lipid, membrane protein, PA phosphatase
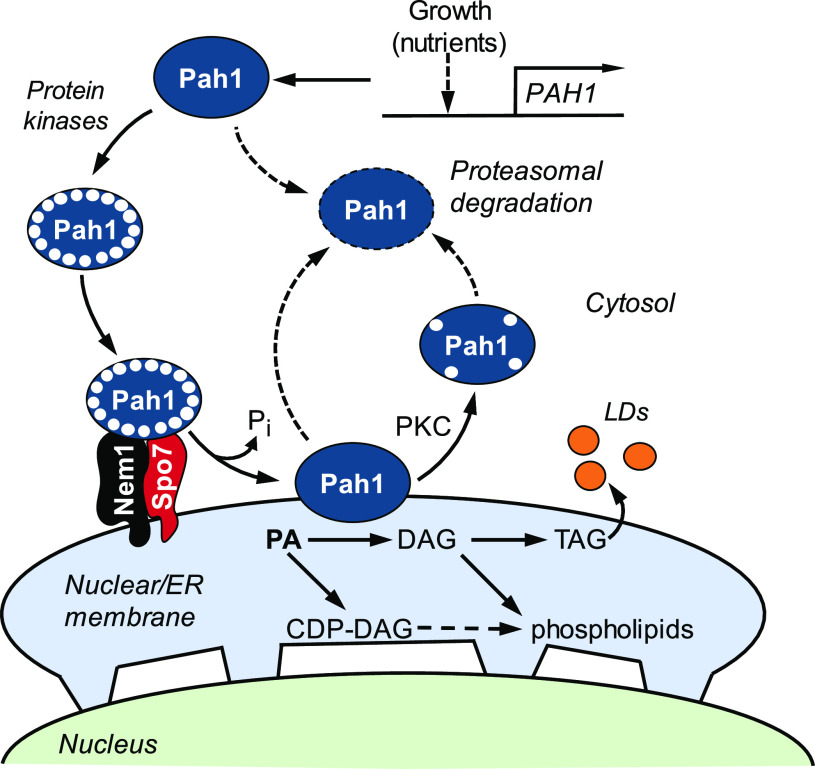
In the model eukaryote Saccharomyces cerevisiae (in this paper, “yeast” is used interchangeably with Saccharomyces cerevisiae), the lipid intermediate phosphatidic acid (PA) is used for the synthesis of phospholipids and the neutral lipid triacylglycerol (TAG) (1–4) (Fig. 1). The bifurcation of PA to these lipids is governed by the demands of cell growth and metabolism (1–4). For example, PA is primarily metabolized into phospholipids via CDP-diacylglycerol (CDP-DAG) (1–4) during logarithmic growth when phospholipids are needed for membrane synthesis, metabolic processe, and cellular signaling (Fig. 1). However, as cells progress into stasis (e.g. stationary phase), PA is primarily metabolized into TAG via diacylglycerol (DAG) (1–4). (Mutants defective in the CDP-DAG-dependent synthesis of phosphatidylcholine and/or phosphatidylethanolamine may synthesize these phospholipids from DAG via the CDP-choline and/or CDP-ethanolamine branches of the Kennedy pathway when supplemented with choline and/or ethanolamine [5–7].) This metabolic switch is largely controlled by the Nem1-Spo7/Pah1 phosphatase cascade (1–4, 8, 9) (Fig. 1).
Figure 1.
Model for Pah1 localization and stability regulated by phosphorylation and dephosphorylation and for its role in lipid synthesis. The expression of PAH1 is regulated during growth by nutrient status as mediated by the Opi1/Ino2-Ino4 (Henry) regulatory circuit and transcription factors Gis1, Rph1, and Zap1 (48, 109). Pah1 is phosphorylated (small white circles) by multiple protein kinases in the cytosol and translocates to the nuclear/ER membrane through its dephosphorylation by the Nem1-Spo7 complex (31, 32). Dephosphorylated Pah1 that is associated with the nuclear/ER membrane via its amphipathic helix (32) catalyzes the conversion of PA to DAG (10), which is converted to TAG for storage in lipid droplets (LDs). The DAG is also used for the synthesis of phosphatidylcholine or phosphatidylethanolamine via the Kennedy pathway when cells are supplemented with choline or ethanolamine (7, 110). Unphosphorylated/dephosphorylated Pah1 or protein kinase C (PKC)-phosphorylated Pah1 (16) is degraded by the 20S proteasome (indicated by the dashed-line arrows) (36).
Pah1 has the molecular function of PA phosphatase (10), which produces DAG by the dephosphorylation of PA (11, 12) (Fig. 1). The enzyme is a phosphoprotein in the cytosol, with its phosphorylation catalyzed by multiple protein kinases (13–18) on at least 40 serine/threonine residues (19–30) (Fig. 2). Overall, the phosphorylation of Pah1 causes its localization in the cytosol and prevents its access to the endoplasmic reticulum (ER) membrane-associated substrate PA, and has the effect of reducing its catalytic activity (13–16, 20). The ER membrane-resident protein phosphatase complex (31), consisting of Nem1 (catalytic subunit) and Spo7 (regulatory subunit), recruits and dephosphorylates Pah1 at the membrane surface and stimulates its PA phosphatase activity (20, 32–34) (Fig. 1). Compared with unphosphorylated/dephosphorylated Pah1, its phosphor-ylated form is more stable against degradation by the 20S proteasome (35, 36), except that it becomes more sensitive to the proteasomal degradation upon phosphorylation by protein kinase C (16). Like its substrate, Pah1, the Nem1-Spo7 phosphatase complex is regulated by phosphorylation (30, 37–41) (Fig. 2). For example, its phosphorylation by protein kinases A (40) and C (41) affects the function of the Nem1-Spo7/Pah1 axis for the decrease and increase, respectively, of TAG synthesis.
Figure 2.
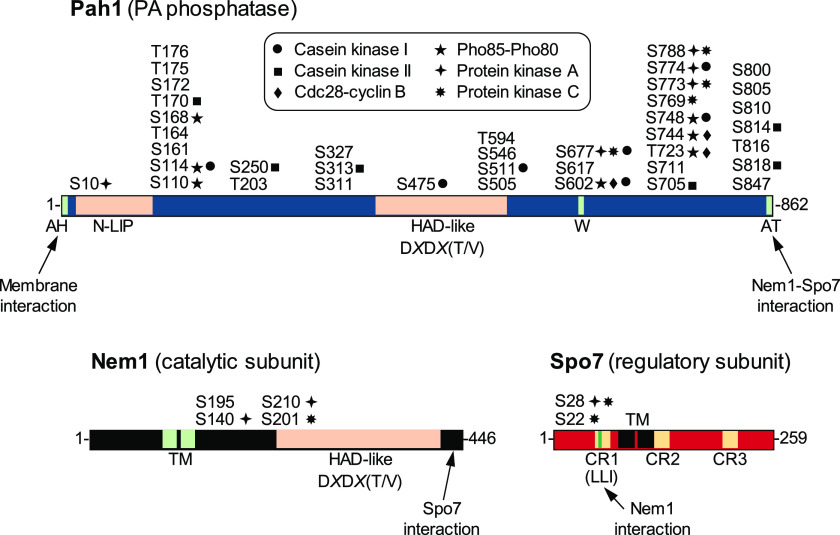
Schematic diagrams for the domains/regions and phosphorylation sites of Pah1, Nem1, and Spo7. The diagram of Pah1 denotes the positions of the amphipathic helix (AH) required for membrane interaction (32), the N-LIP and HAD-like domains required for PA phosphatase activity (42), the tryptophan (W) residue within the C-terminal conserved sequence, WRDPLVDID, required for function in vivo (44), and the acidic tail (AT) required for interaction with the Nem1-Spo7 complex (33). The serine (S) and threonine (T) residues known to be phosphorylated (19–30) are grouped at their approximate positions and marked for the responsible protein kinases, including casein kinase I (18), casein kinase II (17), Cdc28-cyclin B (13), Pho85-Pho80 (11), protein kinase A (15), and protein kinase C (16). For Nem1, the diagram denotes the HAD-like domain required for protein phosphatase activity and the C-terminal region required for interaction with Spo7 (31). For Spo7, the diagram shows the conserved regions (CR) 1, 2, and 3 (yellow) and the LLI sequence (green) within CR1 required for interaction with Nem1 (this work). The transmembrane (TM) regions of Nem1 and Spo7 (31) and the serine residues phosphorylated by protein kinase A (40) or protein kinase C (41) are also indicated.
The activities of Pah1 PA phosphatase (10, 42) and Nem1 protein phosphatase (31, 43) are both dependent on the halo-acid dehalogenase (HAD)-like domain with the DXDX(T/V) catalytic motif (Fig. 2). For Pah1, in addition to the catalytic domain, its N-LIP domain is also important for PA phosphatase activity (42). Pah1 is a peripheral membrane protein that translocates to the ER membrane via its dephosphorylation by the Nem1-Spo7 complex. For Pah1 translocation, its acidic tail at the C terminus is required to interact with the Nem1-Spo7 phosphatase complex (33), whereas its amphipathic helix at the N terminus is crucial to associate with the membrane surface (32). The tryptophan residue of Pah1 in the conserved sequence WRDPLVDID is essential for its in vivo function but is not required for catalytic activity (44). Nem1 and Spo7, which form a protein phosphatase complex, are integral membrane proteins, both of which contain two transmembrane domains (31). The complex formation of Nem1 with Spo7 occurs through its C-terminal conserved domain (31). The interaction of Spo7 with Pah1 (38) facilitates the formation of an enzyme-substrate complex between Nem1 and Pah1 (33).
In the present work, we examined the structural requirement of Spo7 in regulating the function of the Nem1-Spo7/Pah1 phosphatase cascade. Deletion analysis of Spo7 indicated that its amino acid region 46–65 is required for Nem1-Spo7/Pah1 axis function, and the N-terminal region contains the hydrophobic sequence Leu-Leu-Ile (LLI) (residues 54–56), conserved in fungi. Mutational analysis showed that the overall hydrophobicity of the LLI sequence is required for the complex formation of Spo7 with Nem1, and its defect causes a significant loss of the phosphatase complex-controlled Pah1 function (e.g. TAG synthesis and lipid droplet formation, the nuclear/ER membrane morphology, vacuole fusion, and growth at elevated temperature and on glycerol as a carbon source).
Results
Identification of Spo7 LLI sequence as a functional requirement
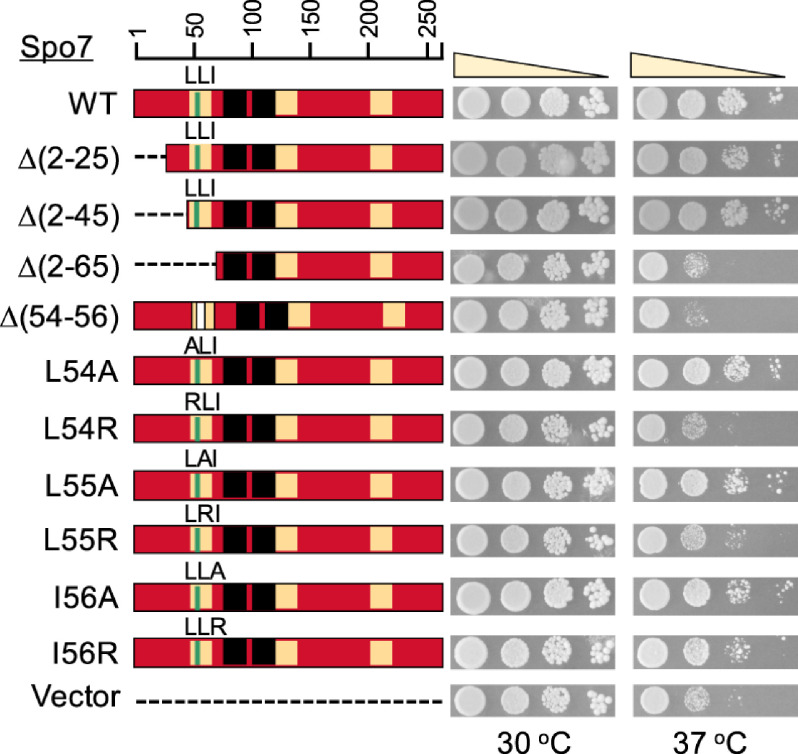
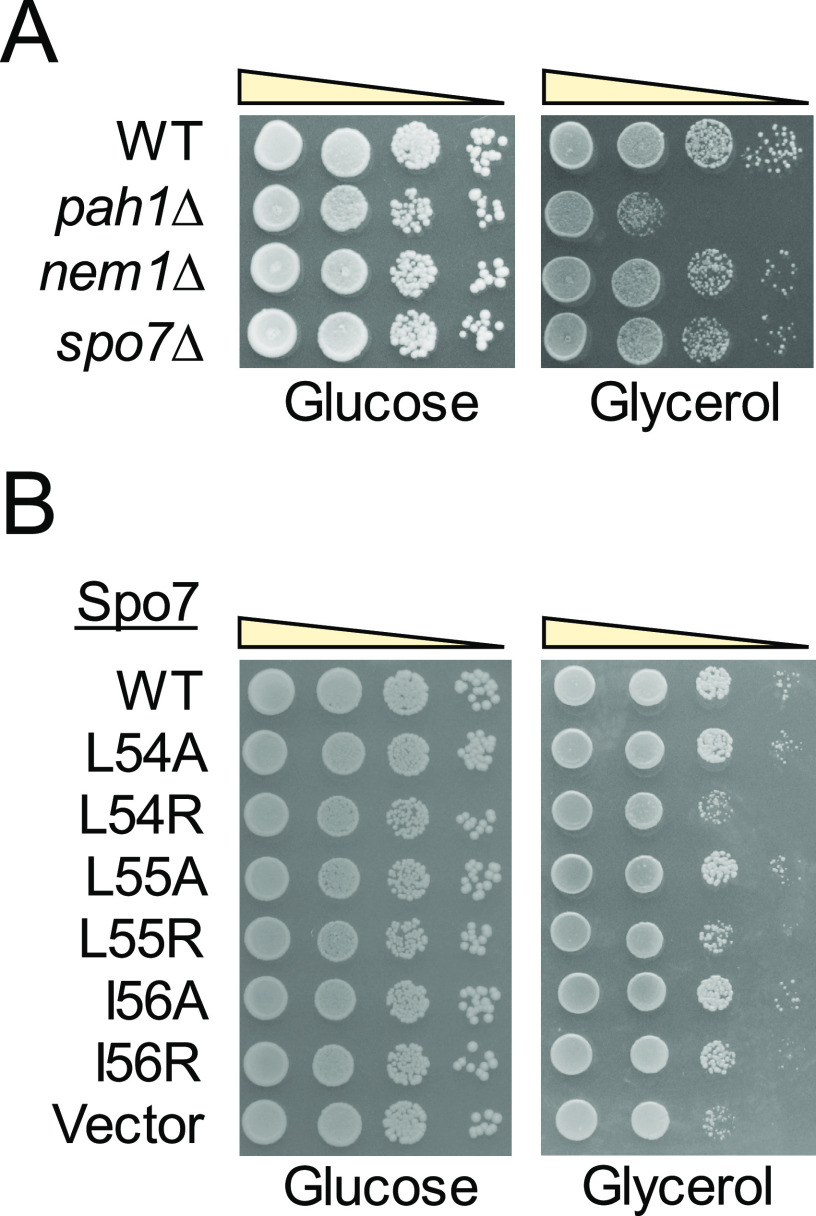
To assess the structural requirement of Spo7 for its function, we mutated the 5′ coding region of SPO7 to produce the nested deletions of the protein from the N terminus (Table 1). The mutant alleles of SPO7 on a low-copy-number plasmid were transformed into the spo7Δ mutant and evaluated for their ability to complement the mutant phenotypes. Loss of Spo7, like that of Pah1 or Nem1, renders cells defective in growth at 37 °C (Fig. 3) (10, 31, 43, 45). Accordingly, we first examined whether the N-terminally truncated forms of Spo7 complement the growth defect of the spo7Δ mutant (Fig. 3). Like the full-length Spo7, its truncation forms Spo7Δ(2–25) and Spo7Δ(2–45) complemented the spo7Δ temperature-sensitive phenotype. However, the expression of Spo7Δ(2–65) did not complement the mutant phenotype. These results indicate that the loss of Spo7 function is caused by the lack of its sequence consisting of amino acids 46 to 65. A protein Blast analysis of the Spo7 sequence revealed the amino acid stretches that are conserved in fungi. We focused on one stretch, namely, LLI (residues 54–56), with the notion that hydrophobic interactions (46) govern the formation of the Nem1-Spo7 phosphatase complex. To examine this possibility, we constructed the SPO7 allele whose product lacks only the LLI sequence. The expression of the LLI-deficient Spo7, Spo7Δ(54–56), did not complement the spo7Δ temperature-sensitive phenotype (Fig. 3), showing that the hydrophobic sequence is required for Spo7 function.
Table 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− ϕ80dlacZΔM15Δ (lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | 95 |
| S. cerevisiae | ||
| RS453 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 | 93 |
| Mutant derivatives | ||
| GHY67 | spo7Δ::URA3 | 40 |
| GHY68 | NEM1-PtA spo7Δ::URA3 | 40 |
| SS1002 | nem1Δ::HIS3 | 31 |
| SS1026 | pah1Δ::TRP1 | 43 |
| Plasmids | ||
| pRS415 | Single-copy E. coli/yeast shuttle vector with LEU2 | 92 |
| pGH443 | SPO7 inserted into pRS415 | 40 |
| pGH443-Δ(2–25) | SPO7 lacking residues 2–25 | This study |
| pGH443-Δ(2–45) | SPO7 lacking residues 2–45 | This study |
| pGH443-Δ(2–65) | SPO7 lacking residues 2–65 | This study |
| pGH443-Δ(54–56) | SPO7 lacking residues 54–56 | This study |
| pGH443-L54A | SPO7 with the L54A mutation | This study |
| pGH443-L54R | SPO7 with the L54R mutation | This study |
| pGH443-L55A | SPO7 with the L55A mutation | This study |
| pGH443-L55R | SPO7 with the L54R mutation | This study |
| pGH443-I56A | SPO7 with the I56A mutation | This study |
| pGH443-I56R | SPO7 with the I56R mutation | This study |
| pRS314-SPO7-Myc | SPO7-Myc inserted into pRS314 | 31 |
| pGH447 | BamH1 site inserted before stop codon of SPO7 in pGH443 | This study |
| pGH448 | 3xMyc tag inserted into SPO7 in pGH447 | This study |
| pGH448-L54A | SPO7-Myc with the L54A mutation | This study |
| pGH448-L54R | SPO7-Myc with the L54R mutation | This study |
| pGH448-L55A | SPO7-Myc with the L55A mutation | This study |
| pGH448-L55R | SPO7-Myc with the L54R mutation | This study |
| pGH448-I56A | SPO7-Myc with the I56A mutation | This study |
| pGH448-I56R | SPO7-Myc with the I56R mutation | This study |
| YCplac33-S.E.C63-GFP | SEC63-GFP fusion inserted into the CEN/URA3 vector | 93 |
| pGH449 | SEC63-GFP fusion inserted into the pRS413 CEN/HIS3 vector | This study |
Figure 3.
Identification of Spo7 LLI sequence as required for the Nem1-Spo7 function. The spo7Δ (GHY68) transformants expressing the indicated SPO7 alleles (pGH443 and its derivatives) were grown to saturation at 30 °C in SC-Leu medium. The cultures were adjusted to an A600 of 0.7, serially diluted (10-fold), and spotted onto SC-Leu agar plates. The growth of the transformant cells on solid medium was scored after 3 days of incubation at 30 and 37 °C. The data are representative of three independent experiments. The position of the amino acid residues from full-length Spo7 is shown (top left).
We next examined the importance of the hydrophobicity of the LLI sequence by mutating each individual amino acid to alanine and arginine. The alanine substitution conserves the hydrophobic property of the leucine and isoleucine residues, whereas the arginine substitution introduces the hydrophilic property instead. The expression of the alanine-substituted forms (i.e. L54A, L55A, and I54A) of Spo7 complemented the spo7Δ temperature-sensitive phenotype (Fig. 3). In contrast, the arginine-substituted forms did not complement (L54R and L55R) or weakly complemented (I56R) the spo7Δ temperature sensitivity (Fig. 3). These results indicate that the hydrophobic property of the LLI sequence is important for Spo7 function, and that the hydrophobic requirement is more stringent for the leucine residues.
Spo7 LLI sequence interacts with Nem1
Spo7 functions as a regulatory subunit of the Nem1-Spo7 phosphatase complex (31, 43). To address the hypothesis that the LLI sequence is required for the protein–protein interaction of Spo7 and Nem1, we examined the mutational effects of the hydrophobic sequence on Nem1-Spo7 complex formation. In this analysis, the isolation of the phosphatase complex and its immunodetection were facilitated by using the protein A-tagged Nem1 and the Myc-tagged Spo7 (20, 31, 34, 40), which are functional to complement the spo7Δ phenotypes (e.g. temperature sensitivity and growth defect on glycerol medium).
The presence of Nem1-PtA in the isolated complex could be determined by immunoblot analysis with anti-protein A or anti-Nem1 antibody. However, anti-protein A antibody was more robust in signal detection; thus, it was used in this analysis. The cross-reactivity of anti-Spo7 antibody with proteins whose electrophoretic mobility overlaps that of untagged Spo7 interfered with the detection of the Spo7-specific signal. To circumvent this problem, we utilized the Myc-tagged version of Spo7 (31) that exhibits slower electrophoretic mobility for clear analysis with anti-Spo7 antibody. Although anti-Myc antibody detected Spo7-Myc, its signal was not as robust as that detected with anti-Spo7 antibody. Nem1 and Spo7 are very-low-abundance proteins (47), and their detection from cell extracts was untenable by immunoblotting with available antibodies. Thus, the typical input (i.e. cell extract) control was not applicable to the Nem1–Spo7 interaction data presented here.
Nem1-PtA was affinity purified with IgG-Sepharose from the extracts of NEM1-PtA spo7Δ cells expressing Spo7-Myc, and its level was determined by immunoblot analysis with anti-protein A antibody (Fig. 4A). In addition, Spo7-Myc copurified with Nem1-PtA was detected by immunoblot analysis with anti-Spo7 antibody (Fig. 4A). The quantification of Spo7-Myc showed that the amounts of the alanine-substituted forms (i.e. L54A, L55A, and I56A) were similar to that of the WT protein (Fig. 4B). However, the levels of the arginine-substituted forms L54R, L55R, and I56R were greatly reduced by 93, 85, and 63%, respectively. This result suggested that the hydrophobicity of the LLI sequence is crucial for Spo7 to form a complex with Nem1. However, we cannot rule out the possibility that the arginine mutations caused reduced stability of the Spo7 protein.
Figure 4.
Nem1-Spo7 complex formation of spo7Δ cells expressing Spo7 with LLI mutations. A, the spo7Δ mutant (GHY68), which harbors chromosomal NEM1-PtA, was transformed with pGH448 and its derivatives for the expression of the WT and mutant forms of Spo7-Myc. The yeast transformants were grown at 30 °C in SC-Leu medium to the late logarithmic phase, and cell extracts were prepared. The cell extracts were adjusted to a protein concentration of 2.5 mg/ml and incubated with IgG-Sepharose. The affinity resins were precipitated, washed, and treated with the Laemmli sample buffer. After centrifugation, the affinity-purified proteins in the supernatant were resolved by SDS-PAGE and transferred to a PVDF membrane. The membrane was cut at the 50-kDa position, and the upper portion was probed with anti-protein A antibody, whereas the lower portion was probed with anti-Spo7 antibody. The positions of Nem1-PtA, Spo7-Myc, and molecular mass standards are indicated. B, the signals of Nem1-PtA and Spo7-Myc in panel A were quantified by ImageQuant software. The levels of mutant Spo7-Myc were normalized to the level (set at 100%) of the WT protein; the levels of Nem1-PtA from the transformant cells expressing mutant Spo7-Myc were normalized to the protein level (set at 100%) from those expressing the WT protein. The density of a background region on the blot was subtracted from the density of the protein band of interest. The immunoblots in panel A are representative of three separate experiments, whereas the data in panel B are averages from the three experiments ± S.D. (error bars). The individual data points are also shown. *, p < 0.05 versus Nem1-PtA of cells expressing WT Spo7-Myc. #, p < 0.05 versus Spo7-Myc of cells expressing WT Spo7-Myc.
For Nem1-PtA, its cellular levels were shown to correlate with the levels of copurified Spo7-Myc (Fig. 4B). Unlike the alanine-substituted forms (i.e. L54A, L55A, and I56A) of Spo7, its arginine-substituted forms, L54R, L55R, and I56R, had the effect of reducing the Nem1-PtA level by 78%, 80%, and 44%, respectively. Considering that the Nem1-PtA level is reduced by 85% in the cell lacking Spo7-Myc expression (i.e. vector control), this result raises the suggestion that Nem1 is unstable when it cannot form a complex with Spo7.
Spo7 LLI sequence is required for the Nem1 catalytic activity
The Nem1-Spo7 phosphatase complex catalyzes the dephosphorylation of native Pah1, which is shown by a small increase in its electrophoretic mobility in SDS-PAGE (13, 14, 20). In contrast, the phosphorylation of Pah1 at Ser-723, Ser-744, and Ser-748 by the Pho85-Pho80 protein kinase is shown by a small decrease in its migration in the polyacrylamide gel (13, 14, 20). The Pho85-Pho80-mediated phosphorylation in the cell is required to ensure that the localization of Pah1 to the membrane depends on its dephosphorylation by the Nem1-Spo7 phosphatase (20). Accordingly, we examined the Nem1-Spo7 phosphatase activity in vivo on the substrate Pah1 using its phosphorylation state, which is reflected by differential electrophoretic mobility (Fig. 5). Compared with the expression of Spo7 (i.e. WT), the lack of its expression (i.e. vector) resulted in the production of the slower-migrating form of Pah1 concomitant with the lack of its faster-migrating form, indicating that the Nem1-Spo7 phosphatase activity is required to dephosphorylate native Pah1.
Figure 5.
Electrophoretic mobility of Pah1 from spo7Δ cells expressing Spo7 with LLI mutations. The spo7Δ mutant (GHY67) was transformed with pGH443 and its derivatives for expression of the WT and mutant forms of Spo7-Myc. The yeast transformants were grown at 30 °C in SC-Leu medium to the mid-logarithmic phase. Cell extracts were prepared and subjected to SDS-PAGE (40 µg protein) using an 8% polyacrylamide gel. A, the proteins resolved in the polyacrylamide gel were transferred to a PVDF membrane and probed with anti-Pah1 antibody. The positions of Pah1 and molecular mass standards are indicated. The white dashed line is a guide to show the range in the electrophoretic mobility of Pah1 from the spo7Δ cells and those expressing WT Spo7. B, the signal intensities of Pah1 along its migration in the region between the dashed white lines in panel A were measured using the line graph function of ImageQuant software. The densitogram of Pah1 from the expression of mutant Spo7 (black line) was compared with those from the presence (green line) and absence (red line) of WT Spo7. The vector control and WT lines are included with each mutant protein for comparison. The data shown are representative of three separate experiments.
The differential electrophoretic mobility of Pah1 was also shown from spo7Δ cells expressing the alanine- and arginine-substituted forms of Spo7 (Fig. 5). The faster-migrating form of Pah1 was shown from the expression of the L54A form, similar to the expression of WT Spo7. In contrast, the slower-migrating form of Pah1 was shown from the expression of the L54R form, similar to the lack of WT Spo7. Compared with the L54A form of Spo7, the L55A and I56A forms showed a less clear effect on producing the faster-migrating form of Pah1. However, the electrophoretic mobilities of Pah1 from the expression of L55A and I56A were shown to be faster than those from the expression of L55R and I56R. The L55R and I56R forms were similar to the L54R form in producing the slower-migrating form of Pah1.
The relative amounts of Pah1 from cells expressing the L54A, L54R, L55A, L55R, I56A, and I56R Spo7 mutant proteins were 92, 100, 87, 91, 99, and 100%, respectively, compared with that of cells expressing WT Spo7 (Fig. 5B). Thus, the Spo7 LLI mutations do not majorly affect the amounts of Pah1.
Mutational effects of Spo7 LLI on the Nem1-Spo7/Pah1 function
Yeast cells deficient in Pah1, Nem1, or Spo7 exhibit a variety of phenotypes, all of which are caused by the lack of Pah1 PA phosphatase activity in producing DAG (4). The most notable phenotypes of the mutants include a great reduction in TAG synthesis (Fig. 6) (10, 48) and lipid droplet formation (Fig. 7A) (49, 50), the aberrant expansion of the nuclear/ER membrane (Fig. 8A) (31, 42, 43), defective vacuole fusion (Fig. 9A) (51–53), and growth defect at elevated temperature (Fig. 3) (10, 43, 45) and on glycerol as a sole carbon source (Fig. 10A) (10, 42). Some of these phenotypes (e.g. aberrant nuclear/ER morphology or defective vacuole fusion) are less pronounced in the spo7Δ mutant than in the pah1Δ mutant (Figs. 8A and 9A) but strong enough to evaluate the mutational effects of the LLI sequence on cell physiology.
Figure 6.
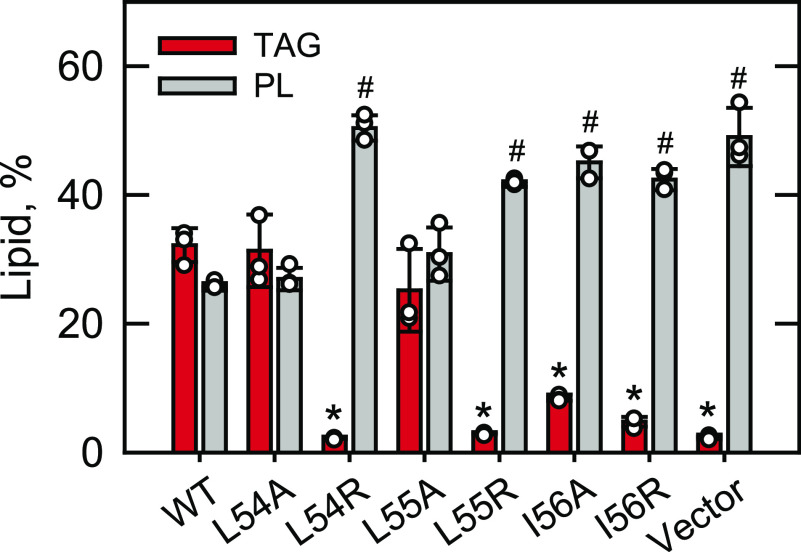
TAG and phospholipid levels of spo7Δ cells expressing Spo7 with LLI mutations. The spo7Δ mutant (GHY68) was transformed with pGH448 and its derivatives for expression of the WT and mutant forms of Spo7-Myc. The transformants were grown at 30 °C to stationary phase in SC-Leu medium containing [2-14C]acetate (1 µCi/ml). Lipids were extracted from the radiolabeled cells, separated by TLC, subjected to phosphorimaging, and quantified by ImageQuant analysis. The levels of TAG and phospholipids (PL) were normalized to total chloroform-soluble lipids. The data are means ± S.D. (error bars) from three separate experiments. The individual data points are also shown. *, p < 0.05 versus TAG of WT. #, p < 0.05 versus phospholipid of WT.
Figure 7.
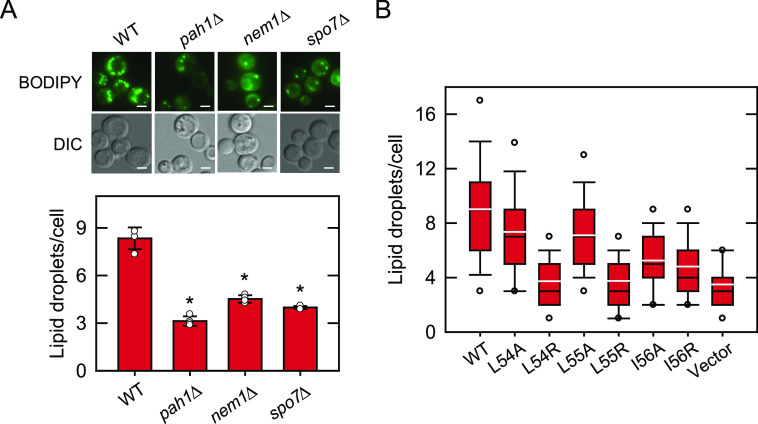
Lipid droplet formation of cells defective in the Nem1-Spo7/Pah1 phosphatase cascade and of spo7Δ cells expressing Spo7 with LLI mutations. A, WT, pah1Δ, nem1Δ, and spo7Δ cells were grown at 30 °C in SC medium to stationary phase and stained with BODIPY 493/503. B, spo7Δ (GHY68) transformants expressing the indicated SPO7 allele (pGH443 and its derivatives) were grown and stained as in panel A, except that SC-Leu medium was used for plasmid selection. The stained lipid droplets were visualized by fluorescence microscopy, and the number of lipid droplets was counted from ≥4 fields of view (≥200 cells). A, upper, the images shown are representative of multiple fields of view. DIC, differential interference contrast. White bar, 2 μm. A, lower, the data shown are averages from three experiments ± S.D. (error bars). The individual data points are also shown. *, p < 0.05 versus cells expressing the WT control. B, the data are presented by the box plot. The black and white lines are the median and mean values, respectively, and the white circles are the outlier data points of the 5th and 95th percentiles.
Figure 8.
Nuclear/ER morphology of cells defective in the Nem1-Spo7/Pah1 phosphatase cascade and of spo7Δ cells expressing Spo7 with LLI mutations. A, WT, pah1Δ, nem1Δ, and spo7Δ cells harboring YCplac111-SEC63-GFP were grown at 30 °C in SC-Leu medium to logarithmic phase, and the fluorescence signal of the GFP-tagged ER marker Sec63 was visualized by fluorescence microscopy. B, spo7Δ (GHY68) transformants harboring pRS413-SEC63-GFP (pGH449) and the indicated SPO7 allele (pGH443 and its derivatives) were grown and examined as in panel A, except that SC-His-Leu medium was used for plasmid selection. The percentage of cells with aberrant nuclear/ER morphology (misshaped versus round nuclei) was determined from ≥4 fields of view (≥200 cells). A, upper, the images shown are representative of multiple fields of view. DIC, differential interference contrast. White bar, 2 μm. A, lower, and B, the data shown are averages from three experiments ± S.D. (error bars). The individual data points are also shown. *, p < 0.05 versus cells expressing the WT control.
Figure 9.
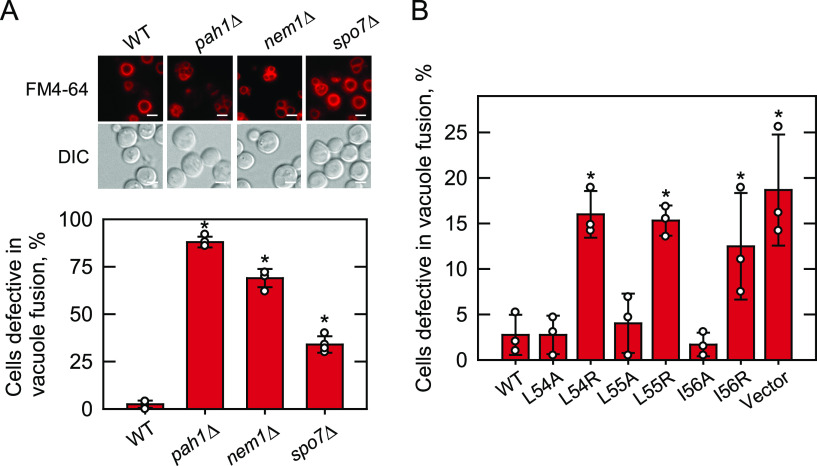
Vacuole morphology of cells defective in the Nem1-Spo7/Pah1 phosphatase cascade and of spo7Δ cells expressing Spo7 with LLI mutations. WT, pah1Δ, nem1Δ, and spo7Δ mutant cells were grown at 30 °C in SC medium to late-logarithmic phase, stained with FM4-64, and examined for vacuole staining by fluorescence microscopy. B, spo7Δ (GHY68) transformants expressing the indicated SPO7 allele (pGH443 and its derivatives) were grown and stained as in panel A, except that SC-Leu medium was used for plasmid selection. The percentage of cells defective in vacuole fusion (e.g. fragmented vacuoles) was determined from ≥4 fields of view (≥200 cells). A, upper, the images shown are representative of multiple fields of view. DIC, differential interference contrast. White bar, 2 μm. A, lower, and B, the data shown are averages from three experiments ± S.D. (error bars). The individual data points are also shown. *, p < 0.05 versus cells expressing the WT control.
Figure 10.
Glycerol utilization of cells defective in the Nem1-Spo7/Pah1 phosphatase cascade and of spo7Δ cells expressing Spo7 with LLI mutations. A, WT, pah1Δ, nem1Δ, and spo7Δ mutant cells were grown at 30 °C in YPD to saturation, serially diluted (10-fold), and spotted onto agar plates containing 2% glucose or 3% glycerol. B, spo7Δ (GHY67) transformants expressing the indicated SPO7 allele (pGH443 and its derivatives) were grown in SC-Leu medium and treated as in panel A. Growth on glucose and glycerol was scored after 3 and 6 days, respectively, of incubation at 30 °C. The data are representative of three independent experiments.
Effect on lipid content
Spo7 is required for the Nem1 catalytic activity, which in turn regulates the Pah1 function to produce DAG for TAG synthesis (10, 20, 32–34, 43, 48). Accordingly, we examined the effect of the site-specific mutations of the LLI sequence on lipid contents (Fig. 6). Lipids were analyzed from the [2-14C]acetate-labeled cells at the stationary phase of growth, when Pah1 PA phosphatase activity is highest (10, 48, 50). Lack of Spo7 (i.e. vector control) caused a 92% decrease in the amount of TAG but a 96% increase in the amount of phospholipids. The altered lipid levels of spo7Δ cells were not restored by expressing the arginine-substituted forms (i.e. L54R, L55R, and I56R) of Spo7. However, the expression of the alanine-substituted forms L54A and L55A restored the altered lipid levels caused by the spo7Δ mutation. Interestingly, the I56A form of Spo7, which complemented the spo7Δ temperature-sensitive phenotype, showed a modest increase in the TAG level without restoring the phospholipid level. The differential effects shown by the I56A form supports the notion that a lower level of Pah1 PAP activity is sufficient to support cell growth at elevated temperature (44).
Effect on lipid droplet formation
Pah1 produces DAG that is acylated to TAG (10), which is then stored in lipid droplets (49). Since the Pah1-controlled production of TAG affects lipid droplet formation, we examined the mutational effects of the LLI sequence on the quantity of cellular lipid droplets (Fig. 7B). The spo7Δ cells expressing WT Spo7 contained an average of 9 lipid droplets in the stationary phase of growth, whereas the mutant cells lacking its expression showed a reduction of the lipid droplet number of 62%. Compared with the effect of the WT control, the arginine-substituted forms (i.e. L54R, L55R, and L56R) of Spo7 did not significantly increase the lipid droplet number of the spo7Δ cells. In contrast, the expression of the alanine-substituted forms L54A and L55A restored the lipid droplet number to 80% of the WT control. Similar to its effect on the TAG level, the I56A form showed a modest increase in the lipid droplet number to 58% of the WT control.
Effect on nuclear/ER morphology
The lack of Spo7, Nem1, or Pah1 in the cell is characterized by an irregularly shaped nucleus with the expansion of the nuclear/ER membrane (Fig. 8A) (31, 42, 43). This phenotype is attributed to the increased synthesis of membrane phospholipids as caused by the defect of the Nem1-Spo7/Pah1 phosphatase cascade (4, 9, 10, 43, 48, 54). Accordingly, we examined the mutational effects of the LLI sequence on the nuclear/ER morphology of the cell by the expression of the ER marker Sec63-GFP (Fig. 8B). On average, about 60% of spo7Δ cells exhibited aberrant nuclear/ER morphology compared with only 3.5% of WT cells. The expression of the alanine-substituted forms of Spo7 significantly reduced the aberrant nuclear/ER morphology of spo7Δ cells (14–22%), whereas the expression of the arginine-substituted forms had weaker effects on suppressing the mutant phenotype (37–42%).
Effect on vacuole fusion
A phenotype characteristic of the cell lacking Pah1, Nem1, or Spo7 is the appearance of fragmented vacuoles that fail to fuse upon nutrient limitation (Fig. 9A) (51, 53). Thus, we examined the effects of the site-specific mutations of the LLI sequence on the vacuole morphology of the cell by staining with FM 4-64. The defect of vacuole fusion was less pronounced in the spo7Δ mutant than in the pah1Δ or nem1Δ mutant (Fig. 9A, lower). As shown in Fig. 9B, about 18% of spo7Δ cells possessed fragmented vacuoles compared with 2.5% of the mutant cells expressing WT Spo7. The expression of the alanine-substituted forms of Spo7 restored the defective vacuole fusion of spo7Δ cells to a level similar to that of the cells expressing the WT protein. In contrast, the expression of the arginine-substituted forms had little effect on restoring the defective vacuole fusion of spo7Δ cells (L54R, 15%; L55R, 15%; I56R, 12%).
Effect on growth with glycerol as the carbon source
Cells lacking Pah1 or Nem1 exhibited a growth defect when glycerol was substituted for glucose as a sole carbon source (Fig. 10A) (42, 45, 55). Because the Nem1-Spo7 complex regulates Pah1 function, we examined whether spo7Δ mutant cells utilize glycerol as a carbon source. The spo7Δ cells exhibited a subtle loss-of-growth phenotype when grown on the glycerol-containing medium; this phenotype was complemented by the expression of WT Spo7 (Fig. 10B). The alanine-substituted forms (i.e. L54A, L55A, and I56A) of Spo7 also complemented the growth defect of spo7Δ cells on glycerol medium. However, the arginine-substituted forms (i.e. L54R, L55R, and I56R) did not complement the spo7Δ defect in glycerol utilization.
Discussion
SPO7, which was originally identified as a gene involved in meiosis and sporulation (56), was shown to encode a protein required to maintain a spherical nuclear morphology (31). The role of Spo7 in sporulation is unclear, but its role in the nuclear morphology is based on the formation of a protein phosphatase complex with Nem1 to regulate Pah1 PA phosphatase (20, 43). The dephosphorylation of Pah1 by the Nem1-Spo7 phosphatase at the nuclear/ER membrane (20, 32, 33) facilitates its membrane localization for catalytic activity on PA to produce DAG that is acylated to TAG (Fig. 1) (48). The storage lipid TAG is a reservoir for energy production as well as for growth resumption from stasis, and its synthesis protects cells against fatty acid-induced toxicity (50, 57–63). TAG production also has the effect of controlling the level of PA that is used for the de novo synthesis of membrane phospholipids. Accordingly, the disruption of the Nem1-Spo7/Pah1 phosphatase cascade by Spo7 deficiency causes the accumulation of PA that is exclusively converted to membrane phospholipids. The elevated level of PA also upregulates the production of phospholipid synthesis enzymes at the transcriptional level through the derepression of enzyme gene expression (10, 43, 48, 50, 54, 64). These changes lead to a massive increase in the level of phospholipids at the nuclear/ER membrane, causing the organelle membrane to be irregularly expanded. The lack of the Spo7-mediated control of the Nem1-Spo7/Pah1 phosphatase cascade also causes defects in other cellular processes (e.g. lipid droplet formation, vacuole fusion, and growth at elevated temperature and on glycerol medium) that are dependent on the regulatory function of PA and/or DAG.
In the Nem1-Spo7 phosphatase complex, Spo7 is a regulatory subunit for the function of the Nem1 catalytic subunit (31, 34, 38, 43, 65) and associates with its HAD-like domain (i.e. catalytic domain) at the C-terminal region (Fig. 2) (31). The work presented here is consistent with the notion that the LLI sequence of Spo7 at the N-terminal region is required for complex formation, and that the overall hydrophobicity of the amino acid residues is crucial for the protein-protein interaction with Nem1. It has been known that residue hydrophobicity is a dominant factor for protein–protein interactions (66). Thus, our findings indicate that the LLI-mediated hydrophobic interaction is a key factor for the formation of the Nem1-Spo7 complex. This mechanism of complex formation could be further supported by the identification of hydrophobic residues in the Nem1 catalytic domain that directly interact with Spo7 LLI. Obviously, the interaction of Spo7 with Nem1 would be better understood from the molecular structural information of the protein complexes. Our results also showed that Nem1 is unstable without forming a complex, indicating that Spo7 is required not only for the function of the catalytic subunit but also for its protein stability. The effect of complex formation on the stability of the catalytic subunit is conserved, as shown by the instability of human CTDNEP1 (catalytic subunit, also called Dullard) in the absence of complex formation with NEP1-R1 (regulatory subunit, also called TMEM188) (65, 67).
The orthologous components of the yeast Nem1-Spo7/Pah1 phosphatase cascade in higher eukaryotes consist of the CTDNEP1–NEP1-R1 complex (65, 67) and lipin PA phosphatase (10, 68–70). Like yeast Pah1, mammalian lipin is controlled for its localization and PA phosphatase activity (71–74) by phosphorylation (75–81) and dephosphorylation (65, 67, 82). In addition, the loss of lipin PA phosphatase is responsible for a variety of lipid-based metabolic disorders. Lipin 1 deficiency in mice and humans causes rhabdomyolysis (83, 84), and its deficiency in mice is also characterized by hepatic steatosis during the neonatal period, lipodystrophy, insulin resistance, and peripheral neuropathy (68, 85). The polymorphism of human LPIN1 is associated with insulin resistance and the metabolic syndrome (86). The deficiency of lipin 2 in humans results in chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anemia (87, 88), whereas the specific polymorphisms of human LPIN2 are associated with type 2 diabetes (89). In mouse small intestine, lipin 2 and lipin 3 deficiencies disrupt phospholipid homeostasis, impairing lipoprotein biogenesis and secretion (90). In mice, the loss of CTDNEP1 causes hemorrhagic ovarian cysts with the accumulation of red blood cells in follicles, leading to infertility (91). Unlike CTDNEP1, NEP1-R1 is not yet known for the effects of its deficiency. Nonetheless, the conservation of the phosphatase cascade indicates that an important avenue of inquiry is to understand how NEP1-R1 interacts with CTDNEP1 for the regulation of lipin activity.
Spo7 is conserved in eukaryotes from fungi to humans (65) by containing two transmembrane-spanning domains (TM1 and TM2) and three homology regions (CR1, CR2, and CR3) (Fig. 2). Whereas the LLI sequence contained within CR1 is conserved in fungi, the two amino acid residues following the first conserved leucine vary in higher eukaryotes (65). Moreover, the second amino acid in the sequence of the worm Caenorhabditis elegans (LKF), the fly Drosophila melanogaster (LKA), and the human Homosapiens (LKA) is the charged amino acid lysine. Thus, based on the data from yeast Spo7, its orthologs containing the lysine residue following the conserved leucine residue are likely to have a weak interaction with the Nem1 orthologs in those organisms.
Compared with CR1 located at the N-terminal region, CR2 and CR3 are located at the C-terminal region. The N-terminal CR1 and the C-terminal CR2/CR3, which are outside the transmembrane regions, are both oriented to the cytosol. The requirement of Spo7 as a regulatory subunit for the catalytic function of Nem1 is thought to be in the recruitment of the substrate. This function of Spo7 requires a region interacting with Pah1, which is separate from a region required to form a complex with Nem1. The LLI sequence of Spo7 located in the N-terminal CR1 is required to form a complex with Nem1. Thus, it is possible that CR2 and CR3 are involved in the recognition of Pah1. In this regard, work is currently in progress to assess the role of CR2 and CR3 in the interaction with Pah1.
Experimental procedures
Materials
Avanti Polar Lipids was the source of lipid standards. Bio-Rad supplied the molecular mass protein standards and reagents for electrophoresis, immunoblotting, and protein determination. DNA size standards were from Invitrogen. Carrier DNA for yeast transformation was from Clontech. Growth media were from Difco Laboratories. GE Healthcare was the source of IgG-Sepharose, polyvinylidene difluoride (PVDF) membrane, and the enhanced chemifluorescence Western blotting detection kit. FM 4-64 was purchased from Molecular Probes, Inc. Millipore-Sigma was the supplier of ampicillin, BSA, 2-mercaptoethanol, PCR primers, nucleotides, Ponceau S stain, Triton X-100, protease and phosphatase inhibitors, rabbit anti-protein A antibody (product P3775, lot 025K4777), rabbit anti-myc antibody (product no. SAB4301136, lot no. 802535536), and silica gel 60 TLC plates. Enzyme reagents for DNA manipulations and the Q5 site-directed mutagenesis kit were obtained from New England Biolabs. Radiochemicals were from Perkin-Elmer Life Sciences, and scintillation-counting supplies were from National Diagnostics. Qiagen was the source of plasmid DNA purification kits. Thermo Scientific was the source of alkaline phosphatase-conjugated goat anti-rabbit IgG antibody (product 31340, lot NJ178812) and BODIPY 493/503. Anti-Nem1 (40), anti-Spo7 (40), and anti-Pah1 (13) antibodies were previously generated in New Zealand White rabbits. All other chemicals were reagent grade.
Strains, plasmids, and DNA manipulations
The strains and plasmids used in this work are listed in Table 1. Escherichia coli strain DH5α was used for plasmid maintenance and amplification. S. cerevisiae strains GHY67 (40) and GHY68 (40), which are spo7Δ mutants with chromosomal NEM1 and NEM1-PtA, respectively, were used for the expression of plasmid-borne SPO7 and its mutant derivatives. Plasmid pGH443 (40), which is a derivative of the E. coli/yeast shuttle vector pRS415 (92), directs the low-copy-number expression of SPO7 in S. cerevisiae. Plasmid pGH447 was derived from pGH443 by introducing a BamHI site before the stop codon of SPO7. pGH448 was constructed from pGH447 by inserting a 126-bp BamHI fragment for the 3×Myc tag that was released from plasmid pRS314-SPO7-Myc (31). The derivatives of plasmids pGH443 and pGH448 with SPO7 mutations were constructed by the Q5 site-directed mutagenesis kit with appropriate primers designed using the NEBaseChanger online software. pGH449 was constructed by ligation of the SmaI/SacI-digested pRS413 (92) with the SEC63-GFP DNA that was released from YCplac33-SEC63-GFP (93) by digestion with HindIII, treatment with Klenow fragment, and digestion with SacI. Standard methods were used for the isolation of chromosomal and plasmid DNA, for the digestion and ligation of DNA, and for the PCR amplification of DNA (94–96). Plasmid transformations of E. coli (95) and yeast (97) were performed as described previously. All gene mutations were confirmed by DNA sequencing.
Growth conditions
Bacterial cells were grown at 37 °C in lysogeny broth medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.4). E. coli transformants containing plasmids were selected by antibiotic (e.g. 100 µg/ml ampicillin) resistance. Standard methods were used for culturing yeast (94, 95). Yeast cells were routinely grown at 30 °C in rich or synthetic complete medium, and those containing plasmids were maintained in synthetic dropout medium. Unless otherwise indicated, 2% glucose was included as a carbon source in growth medium. Solid media for the growth of E. coli and yeast contained agar at a concentration of 1.5 and 2%, respectively. For the measurement of growth on solid medium, serially diluted (10-fold) cultures were spotted onto agar medium, and cell growth was scored after incubation for 3 days at 30 or 37 °C (for temperature sensitivity). Growth on agar medium containing glycerol as the carbon source was scored after incubation for 6 days. Cell density in liquid cultures was estimated spectrophotometrically by measuring absorbance at 600 nm.
Preparation of cell extracts
All steps to prepare cell extracts were performed at 4 °C. Yeast cultures were harvested by centrifugation at 1,500 × g for 5 min. The cells were washed with water and resuspended in lysis buffer (50 mm Tris-HCl [pH 7.5], 0.3 M sucrose, 10 mm 2-mercaptoethanol) containing protease inhibitors (0.5 mm phenylmethylsulfonyl fluoride, 1 mm benzamidine, 5 µg/ml aprotinin, 5 µg/ml leupeptin, and 5 µg/ml pepstatin) and phosphatase inhibitors (2 mm imidazole, 1 mm β-glycerophosphate, 1 mm sodium pyrophosphate, 1 mm sodium fluoride, and 1 mm sodium orthovanadate). The suspended cells were mixed with glass beads (0.5-mm diameter) and then disrupted by five repeats of a 1-min burst and 2-min cooling using a BioSpec Products Mini-Beadbeater-16 (98). The disrupted cells were centrifuged at 1,500 × g for 10 min to separate cell extracts (supernatant) from unbroken cells and cell debris (pellet). The protein concentration of the cell extracts was determined by the method of Bradford (99) using BSA as a standard.
Isolation of the Nem1-Spo7 complex
The protein A-tagged Nem1-Spo7 complex was isolated from cell extracts (2 mg protein) by incubation overnight with IgG-Sepharose (10%, w/v, slurry). The complex was collected by centrifugation at 1,500 × g for 30 s and washed three times with a modified radioimmune precipitation lysis buffer (50 mm Tris-HCl [pH 8.0], 150 mm NaCl, 1% Triton X-100, and 0.1% SDS) containing protease (0.5 mm phenylmethanesulfonyl fluoride, 1 mm benzamidine, 5 µg/ml aprotinin, 5 µg/ml leupeptin, and 5 µg/ml pepstatin) and phosphatase (10 mm sodium fluoride, 5 mm β-glycerophosphate, and 1 mm sodium vanadate) inhibitors (100). The Nem1 and Spo7 proteins were separated by SDS-PAGE (101) using 12% polyacrylamide gels and transferred to PVDF membrane (102–104).
Immunoblotting
Protein transfer from SDS-polyacrylamide gels to PVDF membranes was monitored by Ponceau S staining. The membrane blots were cut; the upper and lower portions were probed with rabbit anti-Nem1 (1 µg/ml) or rabbit anti-protein A (2 µg/ml) antibodies and with rabbit anti-Spo7 (1 µg/ml) or rabbit anti-Myc (1:1,000 dilution) antibodies, respectively. The immunoblot analysis of Pah1 from cell extracts subjected to SDS-PAGE using 8% polyacrylamide gels was performed with rabbit anti-Pah1 antibody (2 µg/ml). The secondary goat anti-rabbit IgG antibody conjugated with alkaline phosphatase was used at a dilution of 1:5,000. Immune complexes on the PVDF membrane were detected using the enhanced chemifluorescence immunoblotting substrate. Fluorimaging, using a Storm 865 Molecular Imager (GE Healthcare), was used to acquire fluorescence signals from immunoblots, and the intensities of the images were analyzed by ImageQuant TL software (GE Healthcare). A standard curve was used to ensure that the immunoblot signals were in the linear range of detection.
Radiolabeling and analysis of lipids
The labeling of cellular lipids with [2-14C]acetate was performed as described previously (105). Lipids were extracted (106) from the radiolabeled cells and then separated by one-dimensional TLC in the solvent system of hexane-diethyl ether-acetic acid (40:10:1, v/v) (107). The resolved lipids were visualized by phosphorimaging and quantified by ImageQuant TL software. The identity of radiolabeled lipids was confirmed by comparison with the migration of authentic standards visualized by staining with iodine vapor.
Fluorescence microscopy
For the fluorescent staining of lipid droplets, yeast cells were grown at 30 °C in synthetic medium to the stationary phase, incubated for 30 min with 2 μm BODIPY 493/503, washed with PBS (pH 7.4), and resuspended in the same buffer. For the fluorescent labeling of nuclear/ER membrane, yeast cells transformed with the SEC63-GFP plasmid (43) were grown at 30 °C in the selection medium to the logarithmic phase and subjected to microscopic analysis. For the fluorescent staining of vacuole membranes, cells were grown at 30 °C in synthetic medium to the late-logarithmic phase, incubated for 15 min at 30 °C with 32 μm FM 4-64, washed with yeast extract peptone dextrose, and resuspended in fresh growth medium (108). The number of lipid droplets per cell and the percentage of cells with aberrant nuclear/ER morphology (misshaped versus round nuclei) or defective vacuole fusion (e.g. fragmented vacuoles) were scored from ≥4 fields of view (≥200 cells). The green and red fluorescence signals were examined under a Nikon Eclipse Ni-U microscope with the EGFP/FITC/Cy2/AlexaFluor 488 and TRITC/Cy3/TagRFP/AlexaFluor 546 filters, respectively, re-corded by the DS-Qi2 camera, and subjected to imaging analysis with NIS-Elements BR software.
Analysis of data
Microsoft Excel software was used for the statistical analysis of data. p values of <0.05 were taken as significant differences.
Data availability
All data are contained within the manuscript.
Acknowledgments
We thank Joanna M. Kwiatek for helpful discussions during the course of this work.
Author contributions—M. M., G.-S. H., and G. M. C. conceptualization; M. M., P. D., G. J. S., Y. P., G.-S. H., and G. M. C. formal analysis; M. M., P. D., G. J. S., Y. P., G.-S. H., and G. M. C. validation; M. M., P. D., G. J. S., Y. P., and G.-S. H. investigation; M. M., P. D., G. J. S., Y. P., G.-S. H., and G. M. C. visualization; M. M., P. D., G. J. S., Y. P., and G.-S. H. methodology; M. M. writing-original draft; P. D., G. J. S., Y. P., G.-S. H., and G. M. C. writing-review and editing; G. M. C. resources; G. M. C. supervision; G. M. C. funding acquisition; G. M. C. project administration.
Funding and additional information—This work was supported, in whole or in part, by National Institutes of Health grants GM028140, GM050679, and GM136128 from the United States Public Health Service. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- PA
- phosphatidic acid
- DAG
- diacylglycerol
- ER
- endoplasmic reticulum
- TAG
- triacylglycerol
- HAD
- haloacid dehalogenase
- LLI
- Leu-Leu-Ile.
References
- 1. Carman G. M., and Han G.-S. (2009) Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 284, 2593–2597 10.1074/jbc.R800059200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carman G. M., and Han G. S. (2019) Fat-regulating phosphatidic acid phosphatase: a review of its roles and regulation in lipid homeostasis. J. Lipid Res. 60, 2–6 10.1194/jlr.S087452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carman G. M. (2019) Discoveries of the phosphatidate phosphatase genes in yeast. J. Biol. Chem. 294, 1681–1689 10.1074/jbc.TM118.004159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwiatek J. M., Han G. S., and Carman G. M. (2020) Phosphatidate-mediated regulation of lipid synthesis at the nuclear/endoplasmic reticulum membrane. Biochim. Biophys. Acta 1865, 158434 10.1016/j.bbalip.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carman G. M., and Henry S. A. (1989) Phospholipid biosynthesis in yeast. Annu. Rev. Biochem. 58, 635–669 10.1146/annurev.bi.58.070189.003223 [DOI] [PubMed] [Google Scholar]
- 6. Carman G. M., and Henry S. A. (1999) Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38, 361–399 10.1016/S0163-7827(99)00010-7 [DOI] [PubMed] [Google Scholar]
- 7. Henry S. A., Kohlwein S., and Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carman G. M., and Han G.-S. (2006) Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 31, 694–699 10.1016/j.tibs.2006.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pascual F., and Carman G. M. (2013) Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim. Biophys. Acta 1831, 514–522 10.1016/j.bbalip.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han G.-S., Wu W.-I., and Carman G. M. (2006) The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218 10.1074/jbc.M600425200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin Y.-P., and Carman G. M. (1989) Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 264, 8641–8645 [PubMed] [Google Scholar]
- 12. Smith S. W., Weiss S. B., and Kennedy E. P. (1957) The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228, 915–922 [PubMed] [Google Scholar]
- 13. Choi H.-S., Su W.-M., Morgan J. M., Han G.-S., Xu Z., Karanasios E., Siniossoglou S., and Carman G. M. (2011) Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286, 1486–1498 10.1074/jbc.M110.155598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi H.-S., Su W.-M., Han G.-S., Plote D., Xu Z., and Carman G. M. (2012) Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 287, 11290–11301 10.1074/jbc.M112.346023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Su W.-M., Han G.-S., Casciano J., and Carman G. M. (2012) Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 287, 33364–33376 10.1074/jbc.M112.402339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Su W.-M., Han G.-S., and Carman G. M. (2014) Cross-talk phosphorylations by protein kinase C and Pho85p-Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem. 289, 18818–18830 10.1074/jbc.M114.581462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hsieh L.-S., Su W.-M., Han G.-S., and Carman G. M. (2016) Phos-phorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism. J. Biol. Chem. 291, 9974–9990 10.1074/jbc.M116.726588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hassaninasab A., Hsieh L. S., Su W. M., Han G. S., and Carman G. M. (2019) Yck1 casein kinase I regulates the activity and phosphorylation of Pah1 phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 294, 18256–18268 10.1074/jbc.RA119.011314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., and Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 10.1074/mcp.M400219-MCP200 [DOI] [PubMed] [Google Scholar]
- 20. O'Hara L., Han G.-S., Peak-Chew S., Grimsey N., Carman G. M., and Siniossoglou S. (2006) Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281, 34537–34548 10.1074/jbc.M606654200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., Villen J., Elias J. E., and Gygi S. P. (2007) Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 6, 1190–1197 10.1021/pr060559j [DOI] [PubMed] [Google Scholar]
- 22. Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., and Hunt D. F. (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U S A 104, 2193–2198 10.1073/pnas.0607084104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smolka M. B., Albuquerque C. P., Chen S. H., and Zhou H. (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U S A 104, 10364–10369 10.1073/pnas.0701622104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Albuquerque C. P., Smolka M. B., Payne S. H., Bafna V., Eng J., and Zhou H. (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7, 1389–1396 10.1074/mcp.M700468-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soufi B., Kelstrup C. D., Stoehr G., Frohlich F., Walther T. C., and Olsen J. V. (2009) Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol. Biosyst. 5, 1337–1346 10.1039/b902256b [DOI] [PubMed] [Google Scholar]
- 26. Gnad F., de Godoy L. M., Cox J., Neuhauser N., Ren S., Olsen J. V., and Mann M. (2009) High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics 9, 4642–4652 10.1002/pmic.200900144 [DOI] [PubMed] [Google Scholar]
- 27. Helbig A. O., Rosati S., Pijnappel P. W., van B. B., Timmers M. H., Mohammed S., Slijper M., and Heck A. J. (2010) Perturbation of the yeast N-acetyltransferase NatB induces elevation of protein phosphorylation levels. BMC Genomics 11, 685 10.1186/1471-2164-11-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soulard A., Cremonesi A., Moes S., Schutz F., Jeno P., and Hall M. N. (2010) The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21, 3475–3486 10.1091/mbc.E10-03-0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P. G., Gerrits B., Picotti P., Lam H., Vitek O., Brusniak M. Y., Roschitzki B., Zhang C., Shokat K. M., Schlapbach R., et al. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 3, rs4 10.1126/scisignal.2001182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Swaney D. L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N. J., and Villen J. (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., and Hurt E. (1998) A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17, 6449–6464 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karanasios E., Han G.-S., Xu Z., Carman G. M., and Siniossoglou S. (2010) A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. U S A 107, 17539–17544 10.1073/pnas.1007974107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karanasios E., Barbosa A. D., Sembongi H., Mari M., Han G.-S., Reggiori F., Carman G. M., and Siniossoglou S. (2013) Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell 24, 2124–2133 10.1091/mbc.E13-01-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Su W.-M., Han G.-S., and Carman G. M. (2014) Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites. J. Biol. Chem. 289, 34699–34708 10.1074/jbc.M114.614883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pascual F., Hsieh L.-S., Soto-Cardalda A., and Carman G. M. (2014) Yeast Pah1p phosphatidate phosphatase is regulated by proteasome-mediated degradation. J. Biol. Chem. 289, 9811–9822 10.1074/jbc.M114.550103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hsieh L.-S., Su W.-M., Han G.-S., and Carman G. M. (2015) Phos-phorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J. Biol. Chem. 290, 11467–11478 10.1074/jbc.M115.648659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Holt L. J., Tuch B. B., Villen J., Johnson A. D., Gygi S. P., and Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dubots E., Cottier S., Peli-Gulli M. P., Jaquenoud M., Bontron S., Schneiter R., and De Virgilio C. (2014) TORC1 regulates Pah1 phosphatidate phosphatase activity via the Nem1/Spo7 protein phosphatase complex. PLoS. One 9, e104194 10.1371/journal.pone.0104194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dey P., Su W. M., Han G. S., and Carman G. M. (2017) Phosphorylation of lipid metabolic enzymes by yeast Pkc1 protein kinase C requires phosphatidylserine and diacylglycerol. J. Lipid Res. 58, 742–751 10.1194/jlr.M075036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Su W.-M., Han G. S., Dey P., and Carman G. M. (2018) Protein kinase A phosphorylates the Nem1-Spo7 protein phosphatase complex that regulates the phosphorylation state of the phosphatidate phosphatase Pah1 in yeast. J. Biol. Chem. 293, 15801–15814 10.1074/jbc.RA118.005348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dey P., Su W. M., Mirheydari M., Han G. S., and Carman G. M. (2019) Protein kinase C mediates the phosphorylation of the Nem1-Spo7 protein phosphatase complex in yeast. J. Biol. Chem. 294, 15997–16009 10.1074/jbc.RA119.010592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han G.-S., Siniossoglou S., and Carman G. M. (2007) The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282, 37026–37035 10.1074/jbc.M705777200 [DOI] [PubMed] [Google Scholar]
- 43. Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., and Siniossoglou S. (2005) The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park Y., Han G. S., and Carman G. M. (2017) A conserved tryptophan within the WRDPLVDID domain of yeast Pah1 phosphatidate phosphatase is required for its in vivo function in lipid metabolism. J. Biol. Chem. 292, 19580–19589 10.1074/jbc.M117.819375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Irie K., Takase M., Araki H., and Oshima Y. (1993) A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236, 283–288 10.1007/BF00277124 [DOI] [PubMed] [Google Scholar]
- 46. Korn A. P., and Burnett R. M. (1991) Distribution and complementarity of hydropathy in multisubunit proteins. Proteins 9, 37–55 10.1002/prot.340090106 [DOI] [PubMed] [Google Scholar]
- 47. Ghaemmaghami S., Huh W.-K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., and Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- 48. Pascual F., Soto-Cardalda A., and Carman G. M. (2013) PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 288, 35781–35792 10.1074/jbc.M113.525766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., and Goodman J. M. (2011) The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fakas S., Qiu Y., Dixon J. L., Han G.-S., Ruggles K. V., Garbarino J., Sturley S. L., and Carman G. M. (2011) Phosphatidate phosphatase activity plays a key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 286, 29074–29085 10.1074/jbc.M111.258798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sasser T., Qiu Q. S., Karunakaran S., Padolina M., Reyes A., Flood B., Smith S., Gonzales C., and Fratti R. A. (2012) The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem. 287, 2221–2236 10.1074/jbc.M111.317420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu X., and Okamoto K. (2018) The Nem1-Spo7 protein phosphatase complex is required for efficient mitophagy in yeast. Biochem. Biophys. Res. Commun. 496, 51–57 10.1016/j.bbrc.2017.12.163 [DOI] [PubMed] [Google Scholar]
- 53. Rahman M. A., Mostofa M. G., and Ushimaru T. (2018) The Nem1/Spo7-Pah1/lipin axis is required for autophagy induction after TORC1 inactivation. FEBS J. 285, 1840–1860 10.1111/febs.14448 [DOI] [PubMed] [Google Scholar]
- 54. Han G.-S., and Carman G. M. (2017) Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis. J. Biol. Chem. 292, 13230–13242 10.1074/jbc.M117.801720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Park Y., Han G. S., Mileykovskaya E., Garrett T. A., and Carman G. M. (2015) Altered lipid synthesis by lack of yeast Pah1 phosphatidate phosphatase reduces chronological life span. J. Biol. Chem. 290, 25382–25394 10.1074/jbc.M115.680314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Esposito R. E., Frink N., Bernstein P., and Esposito M. S. (1972) The genetic control of sporulation in Saccharomyces. II. Dominance and complementation of mutants of meiosis and spore formation. Mol. Gen. Genet. 114, 241–248 10.1007/BF01788893 [DOI] [PubMed] [Google Scholar]
- 57. Fakas S., Konstantinou C., and Carman G. M. (2011) DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae. J. Biol. Chem. 286, 1464–1474 10.1074/jbc.M110.194308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kurat C. F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., and Kohlwein S. D. (2006) Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 281, 491–500 10.1074/jbc.M508414200 [DOI] [PubMed] [Google Scholar]
- 59. Athenstaedt K., and Daum G. (2003) YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278, 23317–23323 10.1074/jbc.M302577200 [DOI] [PubMed] [Google Scholar]
- 60. Athenstaedt K., and Daum G. (2005) Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J. Biol. Chem. 280, 37301–37309 10.1074/jbc.M507261200 [DOI] [PubMed] [Google Scholar]
- 61. Zanghellini J., Natter K., Jungreuthmayer C., Thalhammer A., Kurat C. F., Gogg-Fassolter G., Kohlwein S. D., and von Grunberg H. H. (2008) Quantitative modeling of triacylglycerol homeostasis in yeast–metabolic requirement for lipolysis to promote membrane lipid synthesis and cellular growth. FEBS J. 275, 5552–5563 10.1111/j.1742-4658.2008.06681.x [DOI] [PubMed] [Google Scholar]
- 62. Rajakumari S., Grillitsch K., and Daum G. (2008) Synthesis and turnover of non-polar lipids in yeast. Prog. Lipid Res. 47, 157–171 10.1016/j.plipres.2008.01.001 [DOI] [PubMed] [Google Scholar]
- 63. Kohlwein S. D. (2010) Triacylglycerol homeostasis: insights from yeast. J. Biol. Chem. 285, 15663–15667 10.1074/jbc.R110.118356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kudo S., Shiino H., Furuta S., and Tamura Y. (2020) Yeast transformation stress, together with loss of Pah1, phosphatidic acid phosphatase, leads to Ty1 retrotransposon insertion into the INO4 gene. FASEB J. 34, 4749–4763 10.1096/fj.201901811RR [DOI] [PubMed] [Google Scholar]
- 65. Han S., Bahmanyar S., Zhang P., Grishin N., Oegema K., Crooke R., Graham M., Reue K., Dixon J. E., and Goodman J. M. (2012) Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J. Biol. Chem. 287, 3123–3137 10.1074/jbc.M111.324350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tsai C. J., Lin S. L., Wolfson H. J., and Nussinov R. (1997) Studies of protein-protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci. 6, 53–64 10.1002/pro.5560060106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim Y., Gentry M. S., Harris T. E., Wiley S. E., Lawrence J. C. Jr., and Dixon J. E. (2007) A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc. Natl. Acad. Sci. U S A 104, 6596–6601 10.1073/pnas.0702099104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Péterfy M., Phan J., Xu P., and Reue K. (2001) Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27, 121–124 10.1038/83685 [DOI] [PubMed] [Google Scholar]
- 69. Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., and Reue K. (2007) Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282, 3450–3457 10.1074/jbc.M610745200 [DOI] [PubMed] [Google Scholar]
- 70. Han G.-S., and Carman G. M. (2010) Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 285, 14628–14638 10.1074/jbc.M110.117747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brindley D. N. (1984) Intracellular translocation of phosphatidate phosphohydrolase and its possible role in the control of glycerolipid synthesis. Prog. Lipid Res. 23, 115–133 10.1016/0163-7827(84)90001-8 [DOI] [PubMed] [Google Scholar]
- 72. Cascales C., Bosca L., Martin A., Brindley D. N., and Cascales M. (1988) Age-related changes in the translocation of phosphatidate phosphatase from the cytosol to microsomal membranes in rat liver. Biochim. Biophys. Acta 963, 384–388 10.1016/0005-2760(88)90305-0 [DOI] [PubMed] [Google Scholar]
- 73. Cascales C., Mangiapane E. H., and Brindley D. N. (1984) Oleic acid promotes the activation and translocation of phosphatidate phosphatase. Biochem. J. 219, 911–916 10.1042/bj2190911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Eaton J. M., Mullins G. R., Brindley D. N., and Harris T. E. (2013) Phosphorylation of lipin 1 and charge on the phosphatidic acid head group control its phosphatidic acid phosphatase activity and membrane association. J. Biol. Chem. 288, 9933–9945 10.1074/jbc.M112.441493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., and Lawrence J. C. Jr. (2007) Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282, 277–286. 10.1074/jbc.M609537200 [DOI] [PubMed] [Google Scholar]
- 76. Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., Balderas E., Guertin D. A., Madden K. L., Carpenter A. E., Finck B. N., and Sabatini D. M. (2011) mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 10.1016/j.cell.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zanivan S., Gnad F., Wickstrom S. A., Geiger T., Macek B., Cox J., Fassler R., and Mann M. (2008) Solid tumor proteome and phosphoproteome analysis by high resolution mass spectrometry. J. Proteome Res. 7, 5314–5326 10.1021/pr800599n [DOI] [PubMed] [Google Scholar]
- 78. Grimsrud P. A., Carson J. J., Hebert A. S., Hubler S. L., Niemi N. M., Bailey D. J., Jochem A., Stapleton D. S., Keller M. P., Westphall M. S., Yandell B. S., Attie A. D., Coon J. J., and Pagliarini D. J. (2012) A quantitative map of the liver mitochondrial phosphoproteome reveals posttranslational control of ketogenesis. Cell Metab. 16, 672–683 10.1016/j.cmet.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stockli J., Yang J. Y., and James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 10.1016/j.cmet.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lundby A., Andersen M. N., Steffensen A. B., Horn H., Kelstrup C. D., Francavilla C., Jensen L. J., Schmitt N., Thomsen M. B., and Olsen J. V. (2013) In vivo phosphoproteomics analysis reveals the cardiac targets of b-adrenergic receptor signaling. Sci. Signal. 6, rs11 10.1126/scisignal.2003506 [DOI] [PubMed] [Google Scholar]
- 81. Hennessy M., Granade M. E., Hassaninasab A., Wang D., Kwiatek J. M., Han G.-S., Harris T. E., and Carman G. M. (2019) Casein kinase II-mediated phosphorylation of lipin 1b phosphatidate phosphatase at Ser-285 and Ser-287 regulates its interaction with 14-3-3b protein. J. Biol. Chem. 294, 2365–2374 10.1074/jbc.RA118.007246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wu R., Garland M., Dunaway-Mariano D., and Allen K. N. (2011) Homo sapiens Dullard protein phosphatase shows a preference for the insulin-dependent phosphorylation site of lipin1. Biochemistry 50, 3045–3047 10.1021/bi200336b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zeharia A., Shaag A., Houtkooper R. H., Hindi T., de L. P., Erez G., Hubert L., Saada A., de K. Y., Eshel G., Vaz F. M., Pines O., and Elpeleg O. (2008) Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 83, 489–494 10.1016/j.ajhg.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang P., Verity M. A., and Reue K. (2014) Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 20, 267–279 10.1016/j.cmet.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nadra K., De Preux Charles A.-S., Medard J.-J., Hendriks W. T., Han G.-S., Gres S., Carman G. M., Saulnier-Blache J.-S., Verheijen M. H. G., and Chrast R. (2008) Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 22, 1647–1661 10.1101/gad.1638008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wiedmann S., Fischer M., Koehler M., Neureuther K., Riegger G., Doering A., Schunkert H., Hengstenberg C., and Baessler A. (2008) Genetic variants within the LPIN1 gene, encoding lipin, are influencing phenotypes of the metabolic syndrome in humans. Diabetes 57, 209–217 10.2337/db07-0083 [DOI] [PubMed] [Google Scholar]
- 87. Ferguson P. J., and El-Shanti H. I. (2007) Autoinflammatory bone disorders. Curr. Opin. Rheumatol. 19, 492–498 10.1097/BOR.0b013e32825f5492 [DOI] [PubMed] [Google Scholar]
- 88. Ferguson P. J., Chen S., Tayeh M. K., Ochoa L., Leal S. M., Pelet A., Munnich A., Lyonnet S., Majeed H. A., and El-Shanti H. (2005) Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J. Med. Genet. 42, 551–557 10.1136/jmg.2005.030759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aulchenko Y. S., Pullen J., Kloosterman W. P., Yazdanpanah M., Hofman A., Vaessen N., Snijders P. J. L. M., Zubakov D., Mackay I., Olavesen M., Sidhu B., Smith V. E., Carey A., Berezikov E., Uitterlinden A. G., et al. (2007) LPIN2 is associated with type 2 diabetes, glucose metabolism and body composition. Diabetes 56, 3020–3026 10.2337/db07-0338 [DOI] [PubMed] [Google Scholar]
- 90. Zhang P., Csaki L. S., Ronquillo E., Baufeld L. J., Lin J. Y., Gutierrez A., Dwyer J. R., Brindley D. N., Fong L. G., Tontonoz P., Young S. G., and Reue K. (2019) Lipin 2/3 phosphatidic acid phosphatases maintain phospholipid homeostasis to regulate chylomicron synthesis. J. Clin. Invest. 129, 281–295 10.1172/JCI122595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hayata T., Chiga M., Ezura Y., Asashima M., Katabuchi H., Nishinakamura R., and Noda M. (2018) Dullard deficiency causes hemorrhage in the adult ovarian follicles. Genes Cells 23, 345–356 10.1111/gtc.12575 [DOI] [PubMed] [Google Scholar]
- 92. Sikorski R. S., and Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Han G.-S., O'Hara L., Carman G. M., and Siniossoglou S. (2008) An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283, 20433–20442 10.1074/jbc.M802903200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rose M. D., Winston F., and Heiter P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 95. Sambrook J., Fritsch E. F., and Maniatis T. (1989) Molecular Cloning, A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 96. Innis M. A., Gelfand D. H. (1990) PCR Protocols: A Guide to Methods and Applications (Innis M. A., Gelfand D. H., Sninsky J. J., and White T. J., eds) pp. 3–12, Academic Press, Inc., San Diego, CA [Google Scholar]
- 97. Ito H., Fukuda Y., Murata K., and Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 10.1128/JB.153.1.163-168.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Carman G. M., and Lin Y.-P. (1991) Phosphatidate phosphatase from yeast. Methods Enzymol. 197, 548–553 10.1016/0076-6879(91)97182-x [DOI] [PubMed] [Google Scholar]
- 99. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 100. Harlow E., and Lane D. (1988) Antibodies. A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 101. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 102. Guengerich F. P., Wang P., and Davidson N. K. (1982) Estimation of isozymes of microsomal cytochrome P-450 in rats, rabbits, and humans using immunochemical staining coupled with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry 21, 1698–1706 10.1021/bi00536a035 [DOI] [PubMed] [Google Scholar]
- 103. Burnette W. (1981) Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112, 195–203 10.1016/0003-2697(81)90281-5 [DOI] [PubMed] [Google Scholar]
- 104. Haid A., and Suissa M. (1983) Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96, 192–205 10.1016/s0076-6879(83)96017-2 [DOI] [PubMed] [Google Scholar]
- 105. Morlock K. R., Lin Y.-P., and Carman G. M. (1988) Regulation of phosphatidate phosphatase activity by inositol in Saccharomyces cerevisiae. J. Bacteriol. 170, 3561–3566 10.1128/jb.170.8.3561-3566.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 107. Henderson R. J., and Tocher D. R. (1992) Lipid Analysis (Hamilton R. J., and Hamilton S., eds) pp. 65–111, IRL Press, New York [Google Scholar]
- 108. Vida T. A., and Emr S. D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Soto-Cardalda A., Fakas S., Pascual F., Choi H. S., and Carman G. M. (2012) Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast. J. Biol. Chem. 287, 968–977 10.1074/jbc.M111.313130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Carman G. M., and Han G.-S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Ann. Rev. Biochem. 80, 859–883 10.1146/annurev-biochem-060409-092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript.