Figure 2.
Identification of the PAPome, the Pool of Ribosome-Bound mRNAs in PAPs from the Dorsal Hippocampus
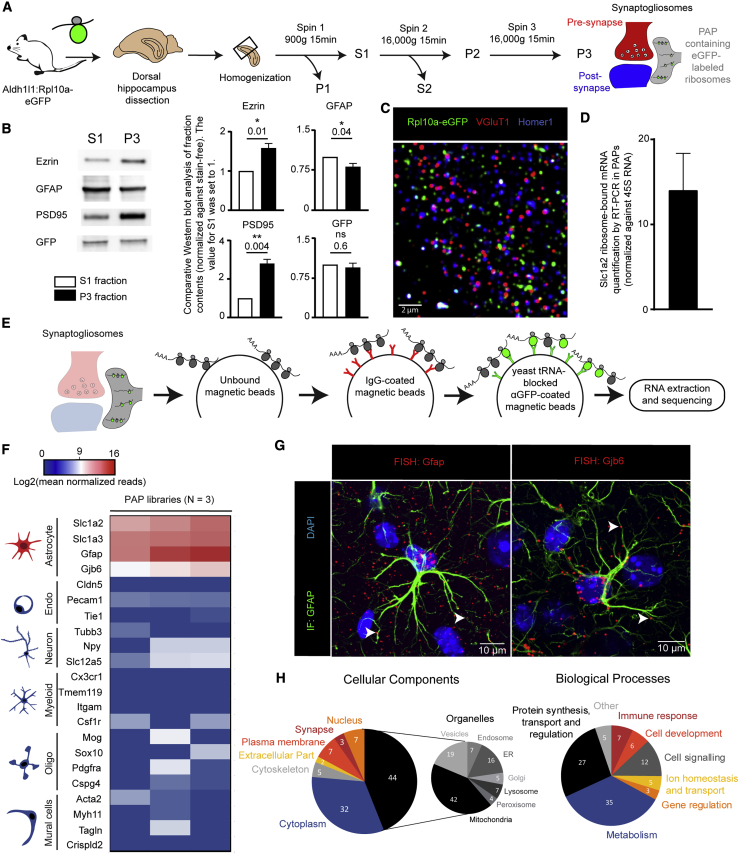
(A) Flowchart for the preparation of synaptogliosomes from the dorsal hippocampus of Aldh1l1:L10a-eGFP transgenic mice. S, supernatant; P, pellet.
(B) Western blot comparison of S1 and P3 fractions. ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ns (not significant) in a two-tailed paired t test. The data are presented as mean ± SEM (n = 4).
(C) A confocal immunofluorescence microscopy image of synaptogliosomes from an Aldh1l1:L10a-eGFP mouse. The ribosomes in PAPs were immunolabeled with eGFP (green). Pre- and postsynaptic areas were immunolabeled for VGluT1 (red) and Homer1 (blue), respectively. Scale bar: 2 µm.
(D) RT-PCR detection of Slc1a2 ribosome-bound mRNAs in synaptogliosomes. The levels were normalized against 45S RNA (n = 4). The data are presented as mean ± SEM.
(E) Flowchart for TRAP and analysis of PAP ribosome-bound mRNAs extracted from dorsal hippocampal synaptogliosomes. The raw sequencing data are given in Table S1.
(F) Purity heatmap of the RNA-seq data for a selection of mRNAs specific for each type of brain cell. The centered value represents mRNAs with log2(500 normalized reads), which corresponds to our threshold for the presence of an mRNA. Each column represents an independent cDNA library (n = 3 libraries).
(G) Confocal microscopy images (FISH detection) of Gfap and Gjb6 mRNAs (red dots) in dorsal hippocampal astrocytes immunolabeled for GFAP (green). Nuclei were stained with DAPI. White arrowheads indicate FISH dots in fine astrocytic processes. Scale bars: 10 µm.
(H) Gene Ontology analysis of the PAPome.