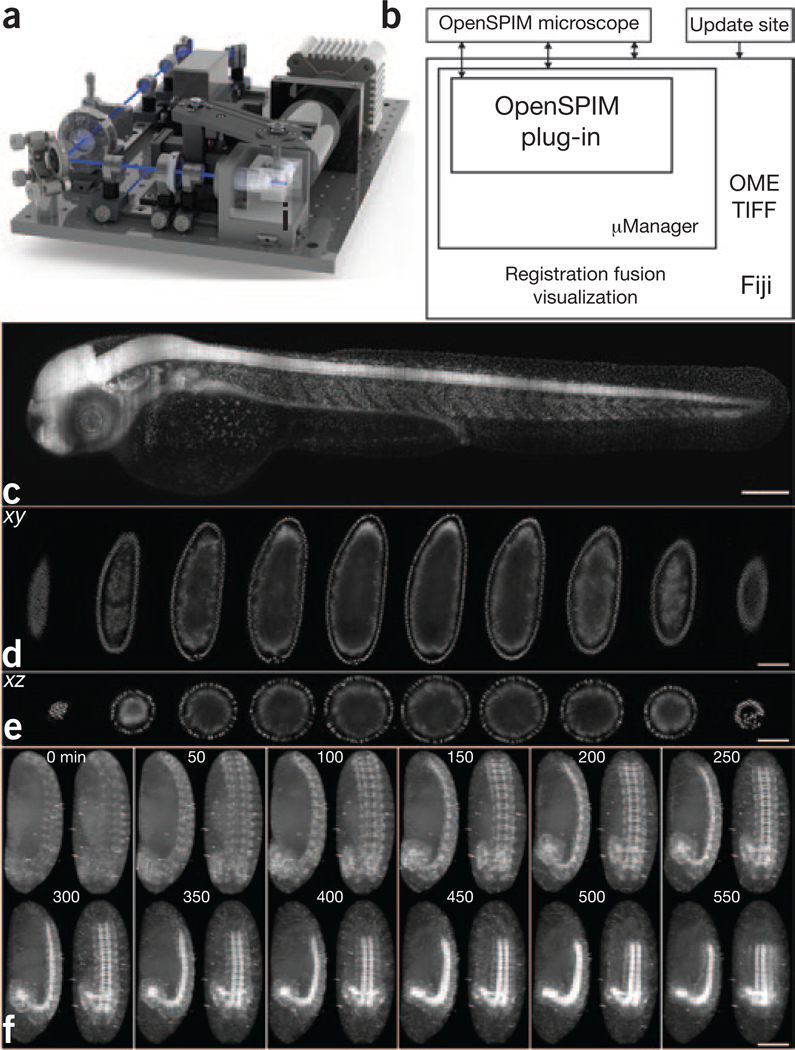
Figure 1 |. OpenSPIM hardware, software and data.
(a) SolidWorks rendering of the OpenSPIM setup. (b) Schematic of the OpenSPIM steering software architecture. (c) A 2-d-old zebrafish larva expressing H2A-EGFP under the control of β-actin promoter Tg(Bactin:H2A-EGFP) was imaged as a set of six overlapping fields of view. (d,e) Lateral (d) and axial (e) planes through a blastoderm-stage Drosophila embryo expressing histone-YFP imaged from six views at 6-μm steps of the light sheet. The data were reconstructed using a bead-based registration algorithm and fused with multiview deconvolution in Fiji. (f) Drosophila embryos expressing Csp-sGFP protein fusion under native promoter control imaged from five views every 10 min. Maximum-intensity projections along the lateral and dorsal-ventral axes are shown for every fifth time point, highlighting the dynamic morphogenetic movement of the nervous system. Scale bars, 100 μm.