Abstract
In the possible event of a detonation of an improvised nuclear device (IND), the immediate radiation would consist of both photons (gamma rays) and neutrons. Since neutrons generally have a high relative biological effectiveness (RBE) for most physiological end points, it is important to understand the effect that neutrons would have on the biodosimetry methods that are being developed for medical triage purposes. We previously compared the transcriptomic response in human blood after neutron and photon irradiation. In this study, we analyzed the effect of mixed-field-neutron-photon radiation on gene expression responses in human peripheral blood, to elucidate the neutron contribution in the setting of a radiation exposure from an IND detonation. We used four combinations of mixed neutronphoton exposures, with increasing percentages of neutrons, to a cumulative dose of 3 Gy. The mixed-field exposures consisted of 0%, 5%, 15% and 25% of neutrons, where 0% corresponds to 3 Gy of pure X rays. A maximum neutron exposure, corresponding to 83% neutrons (0.75 Gy) was also used in the study. Increases were observed in both the number and expression level of genes, with increasing percentages of neutrons from 0% to 25% in the mixed-field exposures. Gene ontology analysis showed an overall predominance of TP53 signaling among upregulated genes across all exposures. Some TP53 regulated genes, such as EDA2R, GDF15 and VWCE, demonstrated increased expression with increasing neutron percentages in mixed-field exposures. Immune response, specifically natural-killer-cell mediated signaling, was the most significant biological process associated with downregulated genes. We observed significant suppression of T-cell-mediated signaling in mixed-field exposures, which was absent in the response to pure photons. In this first study investigating gene expression in human blood cells exposed to mixed neutron-photon fields similar to an actual IND explosion, we have identified a number of genes responding to the 3 Gy dose that showed increasing expression as the neutron percentage increased. Such genes may serve as better indicators of the expected biological damage than a report of total physical dose, and thus provide more relevant information for treating physicians.
INTRODUCTION
While accurate prediction of radiation dose during the first week after a large-scale radiation emergency is a central goal of radiation biodosimetry, most efforts towards developing and validating biomarkers have focused on external gamma- or X-ray exposures (1, 2). In a likely scenario for an improvised nuclear device (IND) detonation, the immediate exposure will be comprised of gamma rays combined with a device-dependent dose of neutrons (3). Since neutrons cause more complex and difficult-to-repair damage to cells that would result in a more severe health burden to affected individuals (4), it is important to understand the effect of neutrons on potential biodosimetry methods. Either an estimate of the neutron contribution or a readout reflective of biological damage, rather than physical dose, will provide important information for those making treatment decisions.
Most of the information on the biological effect of neutrons is based on long-term follow-up studies, such as the Life Span Study (LSS) on atomic bomb survivors in Hiroshima and Nagasaki (5). From the LSS studies, the relative biological effectiveness (RBE) of neutrons for carcinogenesis was estimated as 10 (5). However, there are discrepancies on the magnitude of neutron RBE, which vary based on the study design and biological end points (5–7). The majority of laboratory studies of neutron effects have used fission spectrum neutrons (3, 8). Due to transport through air, however, the neutron spectrum of an IND has a higher proportion of low-energy neutrons, which can alter their biological effect (9, 10). To facilitate studies relevant to IND exposure, our laboratory has developed an IND-spectrum neutron source, with an energy spectrum that simulates exposure at 1.5 km from the site of the Hiroshima blast (11–13).
Gene expression represents an emerging approach to biodosimetry that could provide an estimate of absorbed dose and indicate potential radiation-induced injury (14–19). In previously published studies, we have investigated the effect of neutrons on global gene expression in human blood samples (20) and in mice (21) in comparison with the same dose of photons. In human blood (20), we found a variety of genes that showed similar expression levels after photon and neutron exposures (RBE = 1) as well as genes with a higher-magnitude response to neutrons compared to photons (RBE > 1). The recent mixed-field-neutron-photon study in mice revealed that such exposures resulted in suppression of protein-translational mechanisms mediated through downregulation of ribosomal proteins (22).
In the current study, in support of our development of gene expression signatures for radiation biodosimetry, we irradiated human peripheral blood from healthy donors with graded doses of neutrons using the IND-spectrum neutron irradiator followed by photon irradiation with a cumulative dose of 3 Gy. According to Monte Carlo simulations of an IND explosion at the U.S. Capitol, the neutron contribution of a ground burst in a city area is likely to be much higher than from an airburst, such as that at Hiroshima (9). In the survivable zone, the neutrons are expected to contribute approximately 3–30% of the total physical dose from the IND explosion because a majority of the photons will be shielded by the building structures in the urban area. The maximum neutron dose is expected to be less than 1 Gy outside the blast area, representing the survivable zone that will be relevant for biodosimetry (9, 10). On that basis, in our current study the maximum neutron dose used was 0.75 Gy, representing 25% of a total 3 Gy dose. Human blood samples were exposed to different neutron-photon mixed doses to a total cumulative dose of 3 Gy with increasing percentage of neutron from 5% to 25%, covering the range of neutron contribution expected in the survivable zone. We also used a pure X-ray dose of 3 Gy, and a 0.75 Gy neutron exposure with 0.13 Gy (17%) concomitant gamma rays but no added X rays (83% neutron), for comparison. Sham-irradiated blood samples from the same donors were used as controls. As actual exposure to IND radiation will always be complex and will involve a combination of radiation types, the current study is a step further in our understanding of these complex radiation exposures in human blood.
MATERIALS AND METHODS
Human Donors and Irradiation
Peripheral blood from five healthy human donors was collected in sodium citrate tubes (Becton, Dickinson and Co., Franklin Lakes, NJ). For each donor, one sodium citrate blood tube was exposed to a cumulative dose of 3 Gy using each of four differing percentages (0–25%) of neutrons, as described in Table 1. Another sample was sham irradiated to serve as a control, and one was exposed to 0.75 Gy neutrons (the highest neutron dose used in the 3 Gy study). This sample also received a concomitant 17% gamma dose that is inherent in our source (Table 1). The study was conducted according to the principles in the Declaration of Helsinki and approved by the Columbia University Medical Center Institutional Review Board (IRB no. 4) under protocol IRB-AAAF2671.
TABLE 1.
Different Dose Combinations Used for the Mixed Neutron-Photon Dose Study
| Neutron (%) in mixed neutron-photon irradiation | Neutron dose (Gy) | Photon dose (Gy) | Total dose (Gy) |
|---|---|---|---|
| Sham-control | 0 | 0 | 0 |
| 0% Neutrons (X rays) | - | 3 | 3 |
| 5% Neutrons | 0.15 | 2.84 | 3 |
| 15% Neutrons | 0.45 | 2.47 | 3 |
| 25% Neutrons | 0.75 | 2.12 | 3 |
| 83% (Max.) neutronsa | 0.75 | 0 | 0.75 |
Note. Origianl table published, and modified, with permission, from: Broustas CG, Harken AD, Garty G, Amundson SA. Identification of differentially expressed genes and pathways in mice exposed to mixed field neutron/photon radiation. BMC Genomics 2018; 19:504.
The maximum neutron dose had 17% contaminating photons.
Neutron doses were delivered using a novel neutron irradiator with an energy spectrum based on 1–1.5 km from the epicenter of the Hiroshima blast, to mimic the neutron component expected from IND exposures (12, 13). Blood in citrate tubes was irradiated on a rotating Ferris wheel as described elsewhere (8, 13). Briefly, the tubes were placed in holders designed to provide isotropic exposure as the wheel rotated around the target at approximately one half a revolution per min. Two water-containing tubes were placed before and after the blood-containing tubes to ensure all samples were exposed to the same scatter dose. The tubes were turned end-to-end halfway through the exposure to ensure an equal dose to the whole sample. The dose rate was adjusted to deliver the 0.75 Gy dose in 10 rotations (20 min). Tubes for the different neutron doses were removed from the wheel at specific times according to the dose and replaced with blank tubes containing water. Photon exposures (X rays) were then performed using a Westinghouse Coronado Orthovoltage X-ray machine running at 250 kVp and 15 mA (Pittsburgh, PA).
After irradiation, the blood samples were mixed with an equal volume of RPMI 1640 media supplemented with 10% heat-inactivated fetal bovine serum plus 100 U/ml penicillin and 100 μg/ml streptomycin and incubated for 24 h in a 37°C, 5% CO2 incubator before RNA isolation for gene expression analysis.
Gene Expression Analysis
The effect of mixed neutron-photon exposure was studied on gene expression levels in peripheral blood of healthy individuals using Human Whole Genome Microarrays (4×44K V2; Agilent Technologies Inc., Santa Clara, CA). Total cellular RNA was isolated from blood samples 24 h postirradiation using QIAamp® Blood RNA Mini Kit (QIAGEN®, Valencia, CA). Globin-specific RNA was depleted from total RNA using a GLOBINclear Kit (Thermo Fisher Scientific™ Inc., Waltham, MA). RNA was quantified using the ND-1000 spectrophotometer (NanoDrop, Thermo Fisher Scientific) and quality was assessed using the Agilent Bioanalyzer. Good-quality RNA (100 ng; RIN > 8) was used for Cy3 labeling and amplification using the One-color Low Input Quick Amp Labeling Kit (Agilent Technologies). The labeled RNA were hybridized onto microarrays at 65°C in a hybridization oven (Agilent Technologies) and scanned using the Agilent DNA microarray scanner. The microarray data are available through the NCBI Gene Expression Omnibus (series no. GSE113611; https://bit.ly/2HLiQI8).
Microarrray Data Analysis
The scanned images of the microarrays were used for extracting expression data using Agilent feature extraction software version 10.7. The extracted data were imported into BRB-ArrayTools version 4.3.2 [National Cancer Institute (NCI), Biometric Research Branch, Bethesda, MD] (23) for data filtering, normalization and to identify statistically significant differentially expressed genes. A statistical F test (P < 0.005) at a Benjamini corrected false-discovery rate of <10% was used to identify differentially expressed genes compared to controls (nonirradiated samples).
Gene ontology (GO) and functional analysis for differentially expressed genes for each dose were performed using the Database for Annotation Visualization and Integrated Discovery (DAVID version 6.7) functional annotation tool (24). Benjamini-corrected P value <0.05 was used to identify significant gene ontology terms and biological functions associated with differentially expressed genes.
Ingenuity® Pathways Analysis (IPA®; QIAGEN, http://www.ingenuity.com) was used to identify canonical pathways and predict upstream regulators potentially activated within the gene expression profiles of the different mixed neutron exposures. Statistical z-score (≥2 for activation and ≤−2 for inhibition) was used to identify the predicted activity status of significant canonical pathways and upstream regulators of gene expression. The canonical pathway and upstream regulator analysis specifically uses information about the relationship between the activity of potential upstream regulatory factors or signaling pathways and the expression changes of the measured genes to make predictions of the regulatory status of the upstream molecule or a particular pathway. IPA generates a z-score for each factor in the analysis and for prediction of the activation or inhibition state of each biological function. The IPA default cutoff of z ≥ 2 was used to predict activation and z ≤−2 to predict inhibition.
Quantitative Real-Time RT-PCR
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed for selected genes using TaqMan® chemistry and the Applied Biosystems® 7900 Real-Time PCR System (Carlsbad, CA). The globin-cleared RNA isolated from the irradiated blood of five healthy human volunteers (n = 5) used previously for microarray experiments was used for cDNA synthesis using the High-Capacity cDNA Archive Kit (Life Technologies, Grand Island, NY). Gene expression assays (primer/probe sets) were purchased from Thermo Fisher Scientific for the following genes: GDF15 (Hs00901422_m1), VWCE (Hs00328069_m1), EDA2R (Hs00939736_m1) and ACTB (Hs99999903_m1). The ΔΔCT method was used to calculate expression relative to controls, using normalization to ACTB expression.
RESULTS
Differentially Expressed Genes after Mixed Photon-Neutron Radiation Exposure
We previously reported transcriptomic responses in human blood after neutron exposure at different doses and compared them with the response to photons (X rays) (20). In the current study, we measured the global gene expression response in human blood to mixed neutron-photon irradiations with a cumulative dose of 3 Gy consisting of different percentages of neutrons and photons. The irradiations (Table 1) consisted of a 3 Gy X rays corresponding to 0% neutron, three mixed-field-neutron-photon exposures containing 5, 15 and 25% neutron and a maximum neutron dose corresponding to 83% neutron. Table 2 shows the number of differentially expressed genes (P < 0.005, FDR < 10%) across the different radiation exposures compared to nonirradiated controls (Supplementary Table S1; http://dx.doi.org/10.1667/RR15281.1.S1). We observed an increase in the number of differentially expressed genes with increasing neutron percentage (0–25%) in the 3 Gy mixed-field irradiations. Overall, the number of differentially expressed genes was highest in 25% mixed-neutron exposure, which is the maximum neutron percentage used in the mixed exposures. Fewer genes were affected by the 0.75 Gy (83%) neutron exposure, similar to the number seen in the 0% neutron 3 Gy exposure.
TABLE 2.
Number of Differentially Expressed Genes (BRB Class Comparison; P < 0.005, FDR < 10%) in Response to Different Percentages of Neutrons in a Mixed Neutron-Photon Dose Compared to Controls
| Neutron percentage in mixed neutron-photon irradiation | Number of differentially expressed genes | Number of upregulated genes (percentage of total) | Number of downregulated genes (percentage of total) |
|---|---|---|---|
| 0% Neutrons (X rays) | 385 | 254 (66%) | 131 (34%) |
| 5% Neutrons | 393 | 216 (55%) | 177 (45%) |
| 15% Neutrons | 429 | 262 (61%) | 167 (39%) |
| 25% Neutrons | 689 | 323 (47%) | 366 (53%) |
| 83% (Max.) neutrons | 370 | 192 (52%) | 178 (48%) |
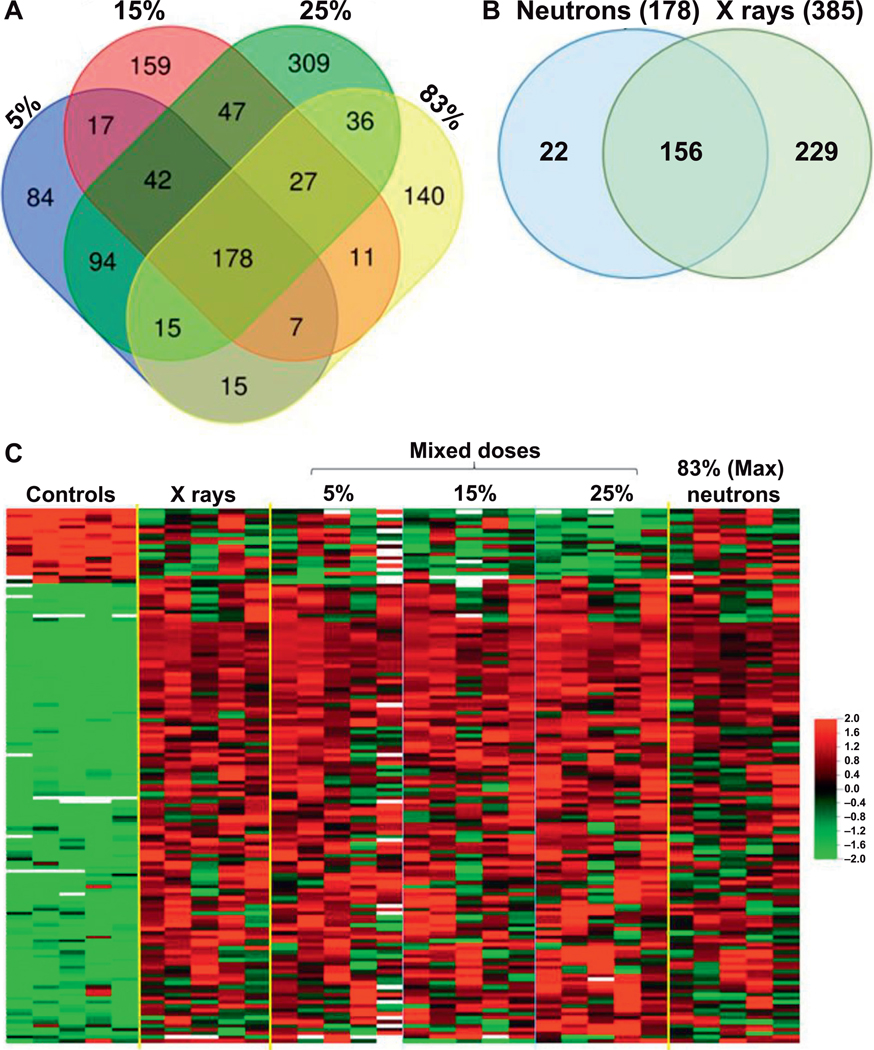
We identified 178 common differentially expressed genes, which responded to radiation containing a neutron component (5%, 15%, 25% and 83% neutrons) (Fig. 1A). Interestingly, 88% (156) of these genes also responded to pure X rays (Fig. 1B). (For Fig. 1A and B, Venn diagrams were generated using the online tool at https://bit.ly/2lZnbti). In a comparison of the genes responding to neutrons beside those responding to pure photons 22 unique genes showed significant expression changes to exposures containing neutrons, but not pure X ray (Fig. 1B; Supplementary Table S1; http://dx.doi.org/10.1667/RR15281.1.S1). Comparative analysis also revealed differential expression of genes that appeared to be unique after exposure to a single neutron percentage, with the largest number responding to the 25% mixed neutron-photon exposure. Further analysis of these gene sets is shown in Supplementary Fig. S1 (http://dx.doi.org/10.1667/RR15281.1.S3).
FIG. 1.
Differentially expressed genes (P < 0.005 and FDR < 10%) Panel A. Venn diagram of differentially expressed genes at different neutron exposures showing overlapping number of genes between different neutron containing exposures (5–83% neutron). In the analysis, 178 common differentially expressed genes were identified across neutron-containing exposures. Panel B. Venn diagram comparing differentially expressed genes in neutron-containing exposures (5–83% neutron) vs. pure X rays (0% neutron). In the analysis, 156 common differentially expressed genes were identified across all exposures. Panel C. Heatmap illustrating expression levels of the common 156 genes across different exposures. “Mixed doses” indicate the percentage neutron exposure in the dose, and “(max). neutron” indicates the exposures of 0.75 Gy neutron, plus 17% contaminating gamma rays. Red indicates high expression and green indicates low expression as shown in the color key. Each row represents one gene and each column represents an individual donor sample.
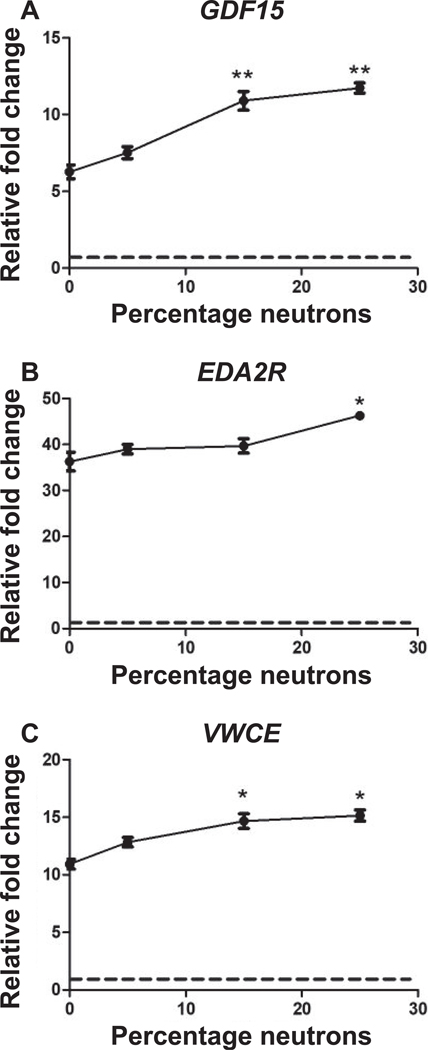
To identify any neutron-specific dose-response effect on gene expression we studied the 156 common genes differentially expressed across all exposures (Fig. 1B). Heatmap visualization of the expression of these common genes showed similarity in expression levels across different percentages of neutron exposures (Fig. 1C). However, in some genes from this set, an increasing trend of expression was evident from 0% to 25% of mixed neutron exposures, with the maximum magnitude of change at 25% neutron (Fig. 1C; Supplementary Fig. S2, http://dx.doi.org/10.1667/RR15281.1.S3). The expression level at 83% neutron was the lowest compared to other mixed neutron exposures (Supplementary Fig. S2). The effect of the neutron component on gene expression in response to mixed-field exposures was more prominent in downregulated genes, which showed a neutron percentage-dependent suppression in expression (Fig. 1C). The expression of several genes (VWCE, GDF15 and EDA2R) was further validated using real-time PCR, which confirmed their increased trend in expression as observed in microarrays (Fig. 2).
FIG. 2.
Real-time quantitative PCR of the genes (panels A–C), GDF15, EDA2R and VWCE, respectively, showing a trend of increased expression in response to a 3 Gy dose with increasing neutron percentages (0% to 25%) in the mixed neutron-photon radiation exposure. The points indicate mean ± SD (n = 5) per exposure condition. **Significant at P < 0.01; *significant at P < 0.05 compared to 0% neutron (X rays) using paired t test analysis. The dashed lines indicate the level in sham-irradiated controls.
Gene Ontology: Functional Analysis of Differentially Expressed Genes
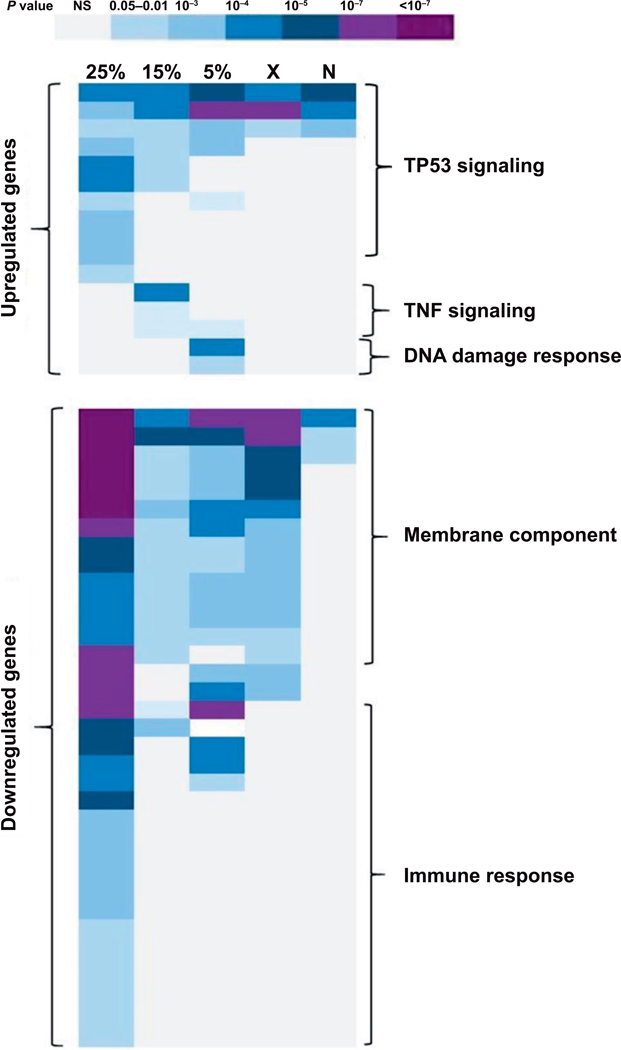
The DAVID functional annotation program was used to find gene ontology terms significantly overrepresented in the transcriptomic responses to different neutron-photon exposures. Similar GO terms were observed to be associated with different neutron exposures (Fig. 3). The TP53 signaling pathway was found to be the top (Benjamini corrected P < 10−7) GO term associated with upregulated genes across all the radiation exposures, whereas immune response and membrane proteins were significantly enriched GO terms in downregulated genes across all exposures. However, we observed enrichment of additional GO terms belonging to TP53 signaling and immune response exclusively in the 25% mixed neutron-photon exposure, and not in other exposures (Supplementary Table S2; http://dx.doi.org/10.1667/RR15281.1.S2). Also, TNF signaling was associated only with 15% neutron irradiation gene expression response, whereas DNA damage was found to be associated specifically with 5% neutron exposure gene expression response (Fig. 3; Supplementary Table S2).
FIG. 3.
Gene ontology (GO) enrichment analysis of differentially expressed genes across all exposures. Using DAVID software (23), the Benjamini corrected P value (P < 0.05) was used for selecting enriched GO categories for each exposure. Darker colors indicate lower P values according to the key. 25% = 25% mixed neutron exposure; 15% = 15% mixed neutron; 5% = 5% mixed neutron; X = 0% neutron (X rays); N = maximum neutron. The predominant biological processes (GO categories) in each region of the heatmap are marked in the figure, with full annotation of individual categories and corresponding P values available in Supplementary Table S2 (http://dx.doi.org/10.1667/RR15281.1.S2).
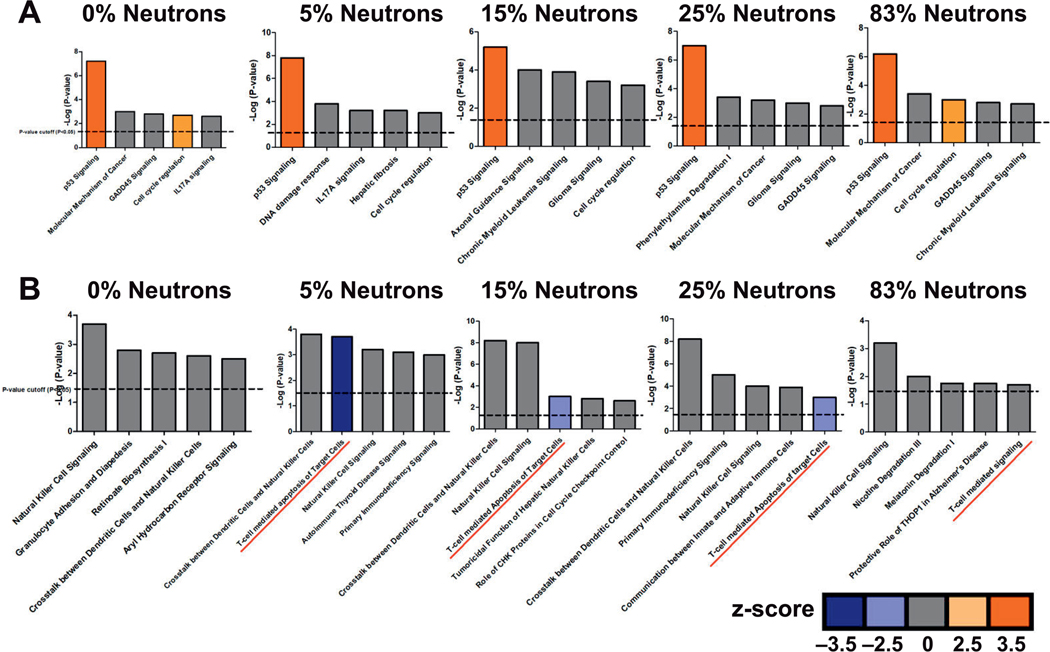
Further analysis was performed using IPA to identify biological processes and pathways significantly (Benjamini corrected P < 0.05) over-represented among differentially expressed genes in different exposure groups. The top five canonical pathways based on the significance of the enrichment P values are shown in Fig. 4. Tp53 was the top pathway associated with upregulated genes across all exposures (Fig. 4A) while immune responses, specifically natural-killer (NK) cell and T-cell-mediated cell signaling, were the top pathways among downregulated genes (Fig. 4B). IPA analysis of unique differentially expressed genes across different neutron exposures also showed a strong association of T-cell-mediated signaling with higher neutron percentage containing mixed-field exposures (Supplementary Fig. S1, http://dx.doi.org/10.1667/RR15281.1.S3).
FIG. 4.
Ingenuity Pathway Analysis (IPA) of the top five canonical pathways (P < 0.05) for each exposure group. The bars represent significant canonical pathways associated with (panel A) upregulated genes and (panel B) downregulated genes in a particular neutron-photon exposure arranged based on their P values (–log; dashed line indicates P = 0.05). The colored bars indicate the predicted activity status of that particular pathway based on statistical z-score (color key). Orange indicates activation (z > 2); blue indicates inhibition (z < −2) and gray indicates no activity pattern available.
Upstream Analysis
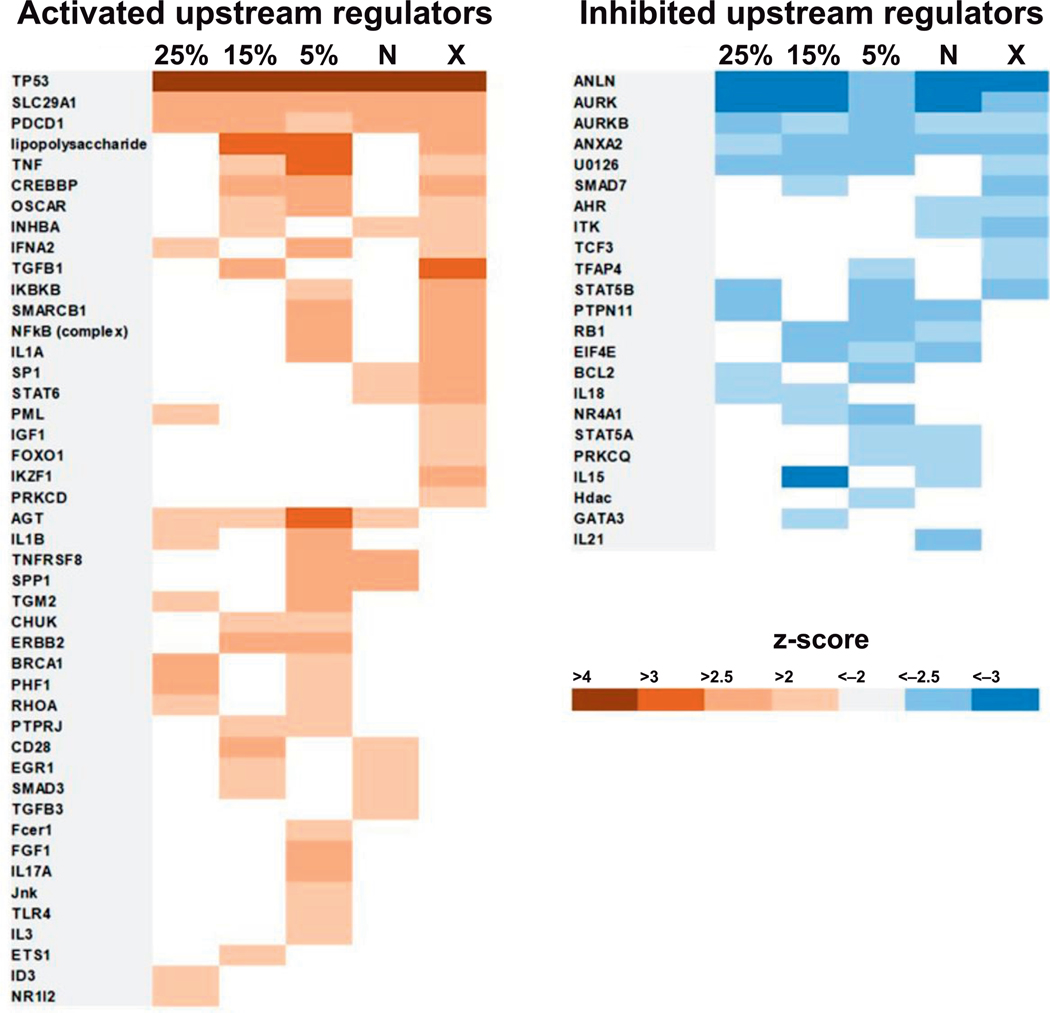
We used IPA analysis to identify upstream regulators in which activity was predicted to be associated with the gene expression profile of a particular neutron exposure group on the basis of statistical z-score. TP53 showed the strongest prediction of activation (z-score > 4) across all irradiation conditions (Fig. 5; Supplementary Table S2, http://dx.doi.org/10.1667/RR15281.1.S2). SLC29A1 and PDCD1 were also predicted to be activated by all the exposures; whereas ANLN, AURK and AURKB were predicted to be suppressed across the study. AGT was predicted to be activated by neutron exposures, but not by pure X rays, while FOXO1, IKZF1 and PRKCD were activated by X rays, but not by exposures including neutrons (Fig. 5). Conversely, TCF3 was predicted to be suppressed specifically in the photon (X ray) only exposure, while IL21 activity was predicted to be suppressed exclusively by the maximum percentage neutron (0.75 Gy) exposure. Overall, the largest number of upstream regulators with significant prediction of activation was associated with the 5% mixed neutron-photon irradiation (Fig. 5; Supplementary Table S2, http://dx.doi.org/10.1667/RR15281.1.S3).
FIG. 5.
Upstream-regulator analysis of differentially expressed genes across all exposures. The IPA tool was used to identify potential upstream regulators of differentially expressed genes. Regulators significant on the basis of z-score (z ≥ 2 or z ≤ −2, P < 0.05) at two or more doses are shown. Positive regulation (activation) is represented in orange and negative regulation (inhibition) is represented in blue. The level of significance of the predicted activity based on statistical z-score is shown in the color key. 25% = 25% mixed neutron exposure; 15% = 15% mixed neutron; 5% = 5% mixed neutron; X = 0% neutron (X rays); N = maximum neutron. The actual z-scores for individual upstream regulators are given in Supplementary Table S2 (http://dx.doi.org/10.1667/RR15281.1.S2).
DISCUSSION
In the current study, we analyzed the transcriptomic response in human peripheral blood to mixed neutron-photon radiation using a neutron energy spectrum similar to that of an IND explosion. As an initial model to study the contribution of neutrons to a mixed-field exposure, we used a fixed cumulative neutron-photon dose of 3 Gy varying the neutron percentage contributing to the total dose.
We previously compared the transcriptomic response of neutrons with similar doses of photons (X rays) and found that many of the same genes respond to both X rays and neutrons but with higher fold change to neutrons compared to X rays (20). In the current study, we focused on identification of genes that were significantly differentially expressed in human blood exposed to a 3 Gy dose of mixed neutron-photon radiation, regardless of the neutron contribution to the total dose. Among these genes, there were some differences in the overall gene expression response to different percentages of neutrons in a 3 Gy total radiation dose (Fig. 1C). We showed that some of these genes, including several identified in our previously reported study, had high neutron RBEs (20), became more highly expressed as the neutron contribution to total dose increased (Fig. 2).
In a previously published study on the effect of mixed-field-neutron-photon exposures on gene expression in mice, we described a large number of eukaryotic translation factors and ribosomal genes differentially expressed 7 days postirradiation including neutrons, but not after X rays alone (22). In the current study, we identified only 22 genes that responded to radiation containing a neutron component, but not to X rays alone (Fig. 1B). The identified genes were mainly involved in transcription regulation [EP300 interacting inhibitor of differentiation 2B (EID2B); E2F transcription factor (E2F7); nuclear factor, erythroid 4 (NFE4); zinc finger and BTB domain, containing 16 (ZBTB16)] and immune response [chemokine (C-X-C motif) ligand 5 (CXCL5); natural cytotoxicity triggering receptor 3 (NCR3); NK cell granule protein 7 (NKG7); killer cell lectin like receptor C1 (KLRC1)]. The differences observed in the two studies could be explained by the different model systems used: mouse in vivo compared with human ex vivo. However, differences in the time of measurement, which in the current study was only 24 h postirradiation, are also likely to play a major role. Neutron damage is more difficult to repair, resulting in greater residual damage than X rays at later times (4, 25). This may result in a greater effect of neutron exposures on gene expression as time progresses, and the X-ray effects diminish as their damage is repaired (22).
In this study, gene ontology analysis revealed that TP53 signaling was the most significant GO term among the upregulated genes across all exposures (Fig. 2; Supplementary Fig. S1; http://dx.doi.org/10.1667/RR15281.1.S3). The dominance of this signaling pathway in the radiation response of human blood has been shown elsewhere (16, 17, 20). However, specific functions associated with this pathway, such as involvement in apoptosis, cell cycle regulation and DNA repair, were over-represented only in the response to the 25% neutron exposure (Fig. 3, Supplementary Table S2, http://dx.doi.org/10.1667/RR15281.1.S2). This result likely reflects the greater damaging effect of the neutron component (highest in 25% neutrons) in the mixed exposures, as was also seen in the greater number of differentially expressed genes at 25% mixed-neutron exposure, despite the fixed dose. We also observed an apparent neutron-dependent increase in gene expression in many TP53-regulated genes (Supplementary Fig. S2). We validated the expression of three of these genes (EDA2R, VWCE and GDF15), which also had among the highest neutron RBEs in our previously reported ex vivo blood study (20), using real-time qRT-PCR. The highest expression was observed in response to 25% neutron exposure (Fig. 2). TP53 is an important transcription factor that gets activated by cellular DNA damage and regulates a variety of cellular functions including apoptosis and DNA repair (26, 27). Published studies have shown that p53 is regulated differently by high-LET and low-LET radiation (28). The more complex cellular damage caused by neutrons at 25% mixed-field exposure could cause a broader spectrum of p53-regulated signaling responses (28, 29) resulting in greater expression of a larger number of p53-regulated genes (Fig. 3).
Among downregulated genes, the analysis revealed significant enrichment of immune response-related biological functions. Detailed analysis of genes involved in immune response revealed that NK cell signaling was most significantly affected in all exposures (Fig. 4). Previously published studies have shown that radiation exposure induces apoptosis in mature NK cells as well as T and B lymphocytes (18, 27, 28). This is consistent with an earlier human ex vivo study (18), in which NK cell-mediated immunity was the most over-represented biological process among downregulated genes 48 h after gamma-radiation exposure, with a simultaneous decrease seen in the percentage of NK cells in the leukocyte population. Individuals exposed to heavy doses of radiation, such as the atomic bomb survivors, also show a profound depletion of granulocytes and NK cells (30).
Our data also indicated significant suppression of T-cell-mediated signaling in exposures containing a neutron component, which was not seen in the transcriptomic response to pure X rays (Fig. 4). To investigate further, we performed an in silico analysis of different blood cell populations across the different exposures using the Cibersort tool (http://cibersort.stanford.edu) (31) to deconvolute the gene expression signatures. In this analysis, we were only able to compare the T-cell population across the different exposures, because the estimated percentage of other cell types in the blood had poor statistical confidence (P > 0.05), and thus did not provide a basis for comparison. This analysis also suggested a significantly decreased T-cell representation in the population after exposures that included any neutron component compared to 3 Gy X-ray irradiation alone (0% neutron) (Supplementary Fig. S3; http://dx.doi.org/10.1667/RR15281.1.S3). Even the 5% neutron exposure, the smallest neutron component tested, appeared to result in the same enhancement of effect on the T-cell population, and the 0.75 Gy neutron dose caused nearly as much decrease as the 3 Gy mixed doses. In vivo mixed-field-neutron-gamma irradiation has previously been shown to cause more prominent and prolonged defects in the T-cell population in mice compared to pure photon irradiation, with effects extending at least 31 days postirradiation (32). This further supports the enhanced effect of neutrons in the mixed-field exposures. We found significant downregulation of T-cell receptor genes as well as perforin and Granzyme B genes after mixed-field exposures but not after pure X ray irradiation, further supporting a major role for T cells in the differential response when neutrons are a part of the exposure.
To look for clues to the regulation of these radiation-responsive genes, we used the gene expression patterns to predict the activation status of upstream regulators. The largest number of regulators was found to be associated with 5% neutron exposure, another indication of the significant effect of minimum neutron percentage in mixed-field exposures on gene expression. While it may seem surprising that the number of regulators predicted to be activated or suppressed did not continue to increase with increasing neutron contribution to the dose, this may be an effect of the increasing numbers of differentially expressed genes. The more diverse response may be spread across more transcription factors, effectively creating more noise, and leading to some of the active regulators no longer appearing significantly affected in the IPA analysis. Major upstream regulators predicted to be activated/inhibited in this study, which are mainly involved in regulation p53 signaling, were consistent with previously reported neutron exposures (20).
In this work, we have demonstrated that a group of genes responds significantly in human blood after a 3 Gy dose that was comprising varying proportions of neutrons in the range that would be relevant for survivors of an IND detonation. Genes showing a greater magnitude of expression response as a function of neutron percentage are strong candidates for use in medical management of radiological casualties, as they may be more reflective of the extent of physiological injury than a physical radiation dose that lacks LET information. Exposures including neutrons appeared to have a greater suppressive effect on T-cell functions than the same dose of X rays alone, suggesting a more profound effect on the hematopoietic system as a precursor of acute radiation syndrome. Future studies will be needed to better understand the relationships between physiological injury, gene expression and exposures to mixed neutrons and photons, as well as other complex radiation exposures.
Supplementary Material
ACKNOWLEDGMENTS
Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team at the NCI. This work was supported by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases (grant no. U19AI067773). Irradiations were performed at the Radiological Research Accelerator Facility (RARAF), an NIH-supported Research Center through NIBIB grant no. 5P41EB-002033.
REFERENCES
- 1.Coleman CN, Koerner JF. Biodosimetry: medicine, science, and systems to support the medical decision-maker following a large scale nuclear or radiation incident. Radiat Prot Dosimetry 2016; 172:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan JM, Prasanna PGS, Grace MB, Wathen LK, Wallace RL, Koerner JF, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys 2013; 105:540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egbert SD, Kerr GD, Cullings HM. DS02 fluence spectra for neutrons and gamma rays at Hiroshima and Nagasaki with fluence-to-kerma coefficients and transmission factors for sample measurements. Radiat Environ Biophys 2007; 46:311–25. [DOI] [PubMed] [Google Scholar]
- 4.Hall EJ, Giaccia AJ. Radiobiology for the radiologist. 7th ed. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2012. [Google Scholar]
- 5.Cullings HM, Pierce DA, Kellerer AM. Accounting for neutron exposure in the Japanese atomic bomb survivors. Radiat Res 2014; 182:587–98. [DOI] [PubMed] [Google Scholar]
- 6.Walsh L. Neutron relative biological effectiveness for solid cancer incidence in the Japanese a-bomb survivors: an analysis considering the degree of independent effects from gamma-ray and neutron absorbed doses with hierarchical partitioning. Radiat Environ Biophys 2013; 52:29–36. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki MS, Endo S, Hoshi M, Nomura T. Neutron relative biological effectiveness in Hiroshima and Nagasaki atomic bomb survivors: a critical review. J Radiat Res 2016; 57:583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young RW, Egbert SD, Cullings HM, Kerr GD, Imanaka T. Survivor dosimetry. Part B. DS02 free-in-air neutron and gamma tissue kerma relative to DS86 In: Young RW, Kerr GD, editors. Reassessment of the atomic bomb radiation dosimetry for Hiroshima and Nagasaki: dosimetry system 2002. Hiroshima, Japan: Radiation effects Research Foundation; (https://bit.ly/2XgG0eQ) [Google Scholar]
- 9.Kramer K, Li A, Madrigal J, Sanchez B, Millage K. Monte Carlo modeling of the initial radiation emitted by a nuclear device in the National Capital Region (Revision 1). Report No. DTRA-TR-13045 (1). Fort Belvoir, VA: Defense Threat Reduction Agency; 2016. [Google Scholar]
- 10.DiCarlo AL, Maher C, Hick JL, Hanfling D, Dainiak N, Chao N, et al. Radiation injury after a nuclear detonation: medical consequences and the need for scarce resources allocation. Disaster Med Public Health Prep 2011; 5:S32–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, Garty G, Marino SA, Massey TN, Randers-Pehrson G, Johnson GW, et al. Novel neutron sources at the Radiological Research Accelerator Facility. J Instrum 2012; 7:C03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Randers-Pehrson G, Turner HC, Marino SA, Geard CR, Brenner DJ, et al. Accelerator-based biological irradiation facility simulating neutron exposure from an improvised nuclear device. Radiat Res 2015; 184:404–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garty G, Xu Y, Elliston C, Marino SA, Randers-Pehrson G, Brenner DJ. Mice and the a-bomb irradiation systems for realistic exposure scenarios. Radiat Res 2017; 187:475–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amundson SA, Bittner M, Meltzer P, Trent J, Fornace AJ. Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat Res 2001; 156:657–61. [DOI] [PubMed] [Google Scholar]
- 15.Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, et al. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med 2007; 4:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol 2008; 71:1236–4.e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul S, Barker CA, Turner HC, McLane A, Wolden SL, Amundson SA. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiat Res 2011; 175:257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul S, Smilenov LB, Amundson SA. Widespread decrease dexpression of immune function genes in human peripheral blood following radiation exposure. Radiat Res 2013; 180:575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghandhi SA, Weber W, Melo D, Doyle-Eisele M, Chowdhury M, Guilmette R, et al. Effect of 90Sr internal emitter on gene expression in mouse blood. BMC Genomics 2015; 16:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broustas CG, Xu, Y, Harken AD, Chowdhury M, Garty G, Amundson SA. Impact of neutron exposure on global gene expression in a human peripheral blood model. Radiat Res 2017; 187:443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broustas CG, Xu Y, Harken AD, Garty G, Amundson SA. Comparison of gene expression response to neutron and x-ray irradiation using mouse blood. BMC Genomics 2017; 3:18:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broustas CG, Harken AD, Garty G, Amundson SA. Identification of differentially expressed genes and pathways in mice exposed to mixed field neutron/photon radiation. BMC Genomics 2018; 19:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-Array Tools. Cancer Inform 2007; 3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 25.Staaf E, Brehwens K, Haghdoost S, Nievaart S, Pachnerova-Brabcova K, Czub J, et al. Micronuclei in human peripheral blood lymphocytes exposed to mixed beams of X-rays and alpha particles. Radiat Environ Biophys 2012; 51:283–93. [DOI] [PubMed] [Google Scholar]
- 26.Nelson WG, Kastan MB. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol 1994; 14:1815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fei P, El-Deiry WS. P53 and radiation responses. Oncogene 2003; 22:5774–83. [DOI] [PubMed] [Google Scholar]
- 28.Niemantsverdriet M, van Goethem M-J, Bron R, Hogewerf W, Brandenburg S, Langendijk JA, et al. High and low LET radiation differentially induce normal tissue damage signals. Int J Radiat Oncol 2012; 83:1291–7. [DOI] [PubMed] [Google Scholar]
- 29.Coelho D, Fischer B, Holl V, Jung GM, Dufour P, Bergerat JP, et al. Involvement of TP53 in apoptosis induced in human lymphoblastoid cells by fast neutrons. Radiat Res 2002; 157:446–52. [DOI] [PubMed] [Google Scholar]
- 30.Kyoizumi S, Kubo Y, Misumi M, Kajimura J, Yoshida K, Hayashi T, et al. Circulating hematopoietic stem and progenitor cells in aging atomic bomb survivors. Radiat Res 2016; 185:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015; 12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cary LH, Noutai D, Salber RE, Williams MS, Ngudiankama BF, Whitnall MH. Interactions between endothelial cells and t cells modulate responses to mixed neutron/gamma radiation. Radiat Res 2014; 181:592–604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.