Figure 1. RREB1; a KRAS-dependent SMAD cofactor.
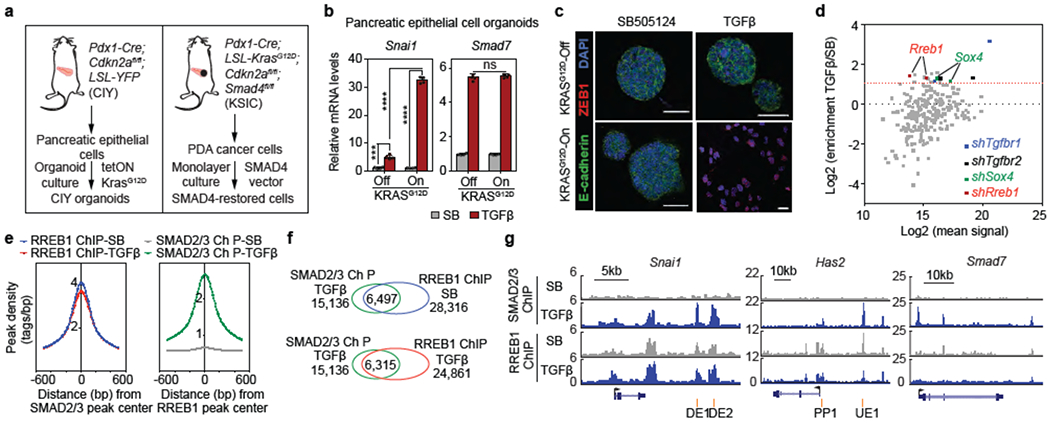
(a) Schematic of source and generation of CIY pancreatic epithelial organoids and SMAD4-restored PDA cells. (b) Snai1 and Smad7 mRNA levels in pancreatic epithelial organoid cultures. Cells engineered to express KRASG12D under doxycycline control treated with TGF^/Nodal receptor inhibitor SB505124 (SB, 2.5 μM) or TGF-β (10 pM) for 1.5 h. Mean ± s.d. n=4, two-way ANOVA analysis, ****, p<0.0001. (c) E-cadherin, ZEB1 and DAPI immunofluorescence images of CIY pancreatic organoids +/− KRASG12D treated with SB or TGF-β for 2.5 days. Scale bars, 30 μm. Images are representative of two independent experiments. (d) Screening of pancreatic progenitor transcription factor shRNA library for mediators of TGF-β-induced lethal EMT. Dot plot of shRNA enrichment in TGF-β-treated versus SB-treated SMAD4-restored PDA cells. Sox412 and Rreb1 transcription factors scoring positive in the screen. shRNAs targeting Tgfbr1 and Tgfbr2 included as positive controls. (e) Position of RREB1 peak summits relative to summits of overlapping SMAD2/3 peaks (left), and position of SMAD2/3 peak summits relative to summits of overlapping RREB1 peaks (right), based on ChIP-seq analysis in Extended Data Figure 2d. (f) Venn diagram depicting overlap between SMAD2/3 and RREB1 ChIP-seq peaks, based on ChIP-seq analysis in Extended Data Figure 2d. (g) Gene track view of SMAD2/3 and HA-RREB1 ChIP-seq tags at indicated loci and experimental conditions. Gene bodies represented at bottom of track sets. PP: proximal promoter; DE: downstream enhancer; UE: upstream enhancer. ChIP-seq was performed once and an independent ChIP was performed in which selective genomic regions were confirmed by quantitative PCR (qPCR). See also Extended Data Figure 1–3 and Supplemental Information Movie 1.
