Figure 4. RREB1 and SMAD regulate distinct context-dependent EMTs.
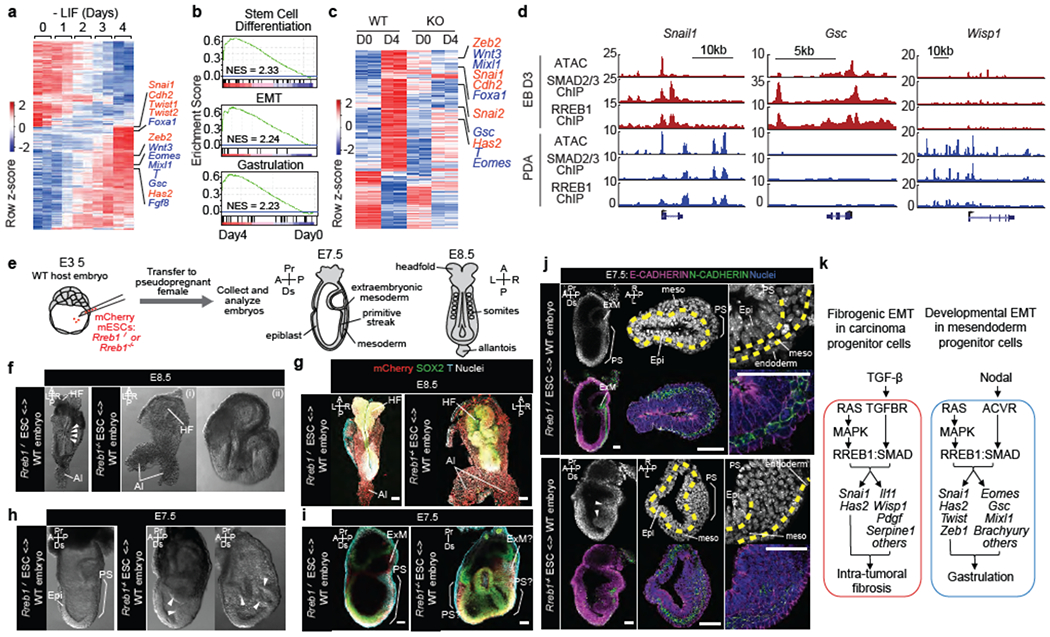
(a) Heatmap representing transcripts up- or down-regulated during embryoid body (EB) differentiation. RNA-seq was performed at indicated times after shifting ESCs into differentiation medium (−LIF). EMT (red) and mesendoderm lineage genes (blue) are highlighted. n=2. (b) Gene set enrichment analysis (GSEA) for EMT, stem cell differentiation and gastrulation signatures in Day 4 EBs. (c) Transcripts exhibiting extensive up- or down-regulation (fold change > 4 or < 0.25) in WT and RREB1 KO cells, on Day4 relative to Day0 of differentiation. n=2. (d) Gene track view of ATAC-seq, and SMAD2/3 and RREB1 ChIP-seq tags at indicated loci, in Day3 EBs (red tracks) versus TGF-β treated (1.5 h) PDA cells (blue tracks). ATAC-seq and ChIP-seq were performed once. Independent ATAC and ChIP were performed in which selected genomic regions were confirmed by qPCR. (e) Chimeras generated by injecting wild-type (WT) Rreb1+/+ or mutant Rreb1−/− mCherry tagged ESCs into WT mouse blastocysts were transferred to pseudopregnant females and dissected at E7.5-E8.5. (f,h) Brightfield images of WT and Rreb1−/− chimeric embryos at E8.5 (f) and E7.5 (h). Rreb1−/− chimeras displayed morphological defects. Arrowheads in (f), somites. Arrowheads in (h), abnormal accumulation of cells within epiblast. (g,i,j) Confocal images of wholemount immunostained chimeras. (g) Maximum intensity projections of E8.5 chimeras. Rreb1−/− chimera shows abnormal neurectoderm development and axis duplication (double allantois). (i) Sagittal section showing Brachyury expression in multiple regions and extensive epiblast folding and multiple cavities in Rreb1−/− chimera (right panel). Due to abnormal morphology, anterior-posterior orientation of the embryo was not possible, (j) Sagittal sections of whole chimeras and representative sections through primitive streak region. Arrowheads, abnormal epiblast folding. Yellow dashed lines, boundary between epiblast and mesoderm. Brackets, primitive streak. HF, headfold; NT, neural tube; Al, allantois; Epi, epiblast; PS, primitive streak; ExM, extraembryonic mesoderm; meso, mesoderm; A, anterior; P, posterior; Pr, proximal; Ds, distal; L, left; R, right. Scale bars, 50 μm. (f-j) Images are representative of two independent experiments. (k) Summary of RAS-dependent TGF-β or Nodal effects, coordinately triggered by cooperation between RREB1 and SMAD2/3 to activate EMT and associated contextual programs in carcinoma progenitors and pluripotent embryonic cells. Principal RREB1/SMAD2/3 target genes in each program and context are indicated. See also Extended Data Figures 8–10.
