Extended Data Figure 2. RREB1 interacts with SMAD and binds to TGF-β target genes.
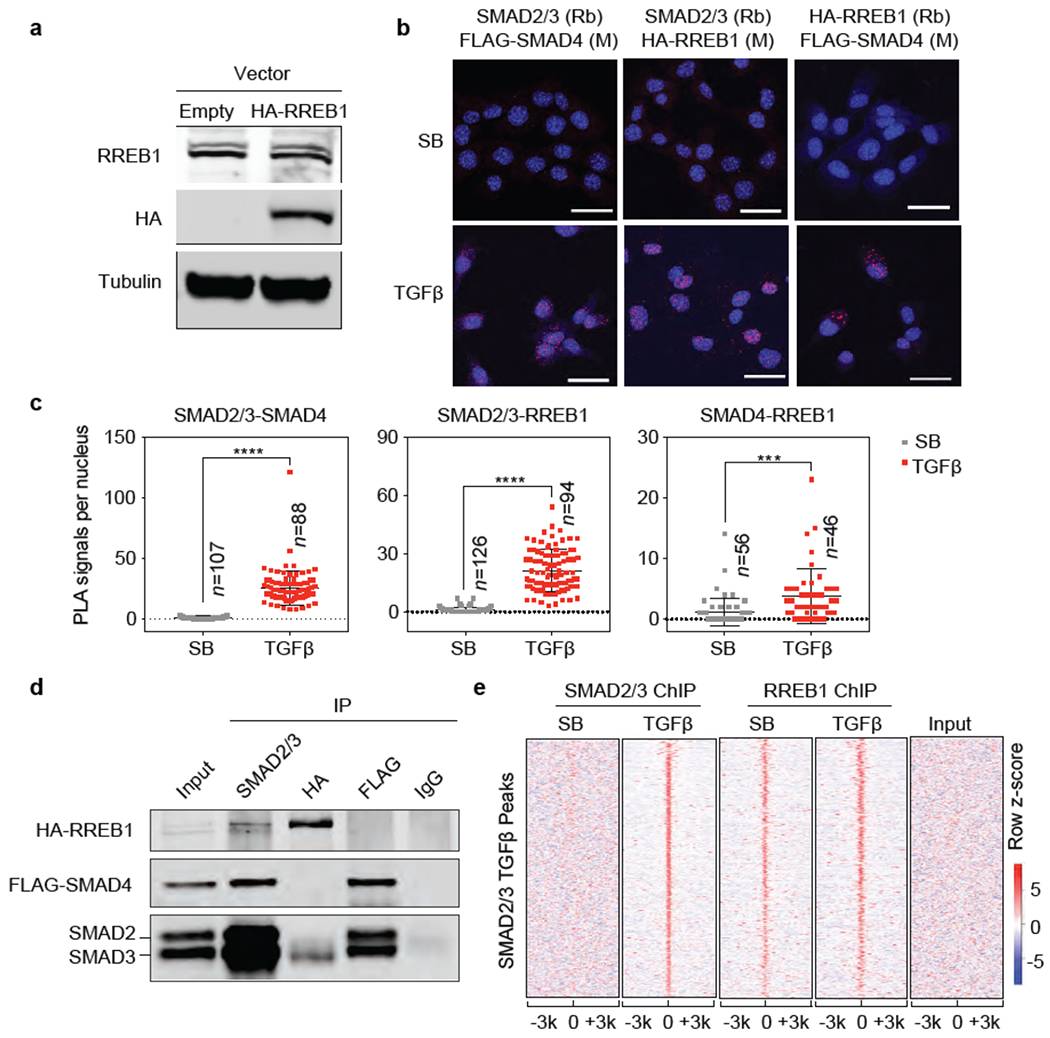
(a) Western immunoblot analysis of RREB1 and HA-RREB1 levels in SMAD4-restored PDA cells stably transduced with an HA-RREB1 vector. Anti-Tubulin immunoblotting was used as loading control. Data are representative of two independent experiments, (b) Proximity ligation assay showing TGF-β dependent proximity between RREB1, SMAD2/3 and SMAD4 in the nucleus. Scale bars, 30 μm. Data are representative of two independent experiments, (c) Quantification of PLA signals in (b). Cell numbers (n) of each group is indicated in the graph, two-tailed unpaired t test. Center values and error bars: mean ± s.d. ****, p<0.0001; ***, p<0.001. (d) SMAD4-restored PDA cells expressing HA-RREB1 were treated with TGF-β for 1.5 h, lysed, and immunoprecipitated (IP) with the indicated antibodies. The immune complexes were collected and subjected to western immunoblot with the antibodies indicated on the left. Data are representative of two independent experiments. (e) Heatmap representation of ChIP-seq tag densities for SMAD2/3 and HA-RREB1 in genomic regions ±3 kb from the center of SMAD2/3 binding peaks in SMAD4-restored PDA cells that were treated with SB or TGF-β for 1.5 h and subjected to SMAD2/3 and HA-RREB1 ChIP-seq analysis. ChIP-seq was performed once and an independent ChIP was performed in which selective genomic regions were confirmed by qPCR.
