Abstract
Aptamers are nucleic acid-based affinity reagents that have gained widespread attention as biorecognition elements for the detection of targets such as ions, small molecules, and proteins. Over the past three decades, the field of aptamer-based sensing has grown considerably. However, the advancement of aptamer-based small-molecule detection has fallen short of the high demand for such sensors in applications such as diagnostics, environmental monitoring, and forensics. This is due to two challenges: the complexity of developing generalized sensing platforms and the poor sensitivities of assays targeting small molecules. This paper will review new approaches for the streamlined development of high-performance aptamer-based sensors for small-molecule detection. We here provide historical context, explore the current state-of-the art, and offer future directions—with emphasis placed on new aptamer engineering methods, the use of cooperative binding, and label-free approaches using fully-folded, high-affinity aptamers for small-molecule sensing.
Keywords: Aptamer, aptamer engineering, biosensor, cooperativity, detection, dye-displacement, enzyme-assisted target recycling, exonuclease, small molecule, sensor
1. Introduction
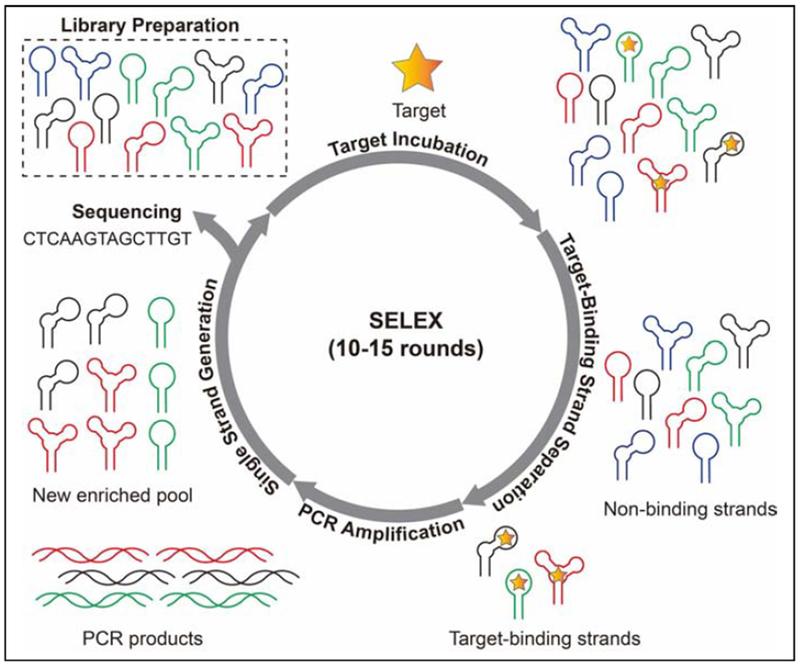
Aptamers are single-stranded oligonucleotides isolated in vitro via a process termed Systematic Evolution of Ligands by Exponential Enrichment (SELEX) to bind to specific molecule [1,2]. SELEX entails challenging a random DNA or RNA library with the target of interest, partitioning target-binding from non-binding strands, amplifying the binders via polymerase chain reaction, and converting the double-stranded amplicons into a new single-stranded pool for the next round. Throughout the isolation process, the molar ratio between the target and library can be adjusted to alter selection stringency to effectively enrich high-affinity aptamers [3]. This process is iterated until the pool is predominated with sequences that bind to the target (Fig. 1). DNA sequencing techniques such as cloning and Sanger sequencing [4] or high throughput sequencing platforms such as Illumina [5] are used to identify the sequence of aptamers. Given that SELEX is entirely done in a test tube, several aspects of the isolation process can be customized. For example, the ionic strength, pH, temperature, and overall chemical environment can be precisely controlled in accordance with the final application of the aptamer. Several different selection strategies have employed during SELEX to isolate aptamers with specific binding properties [6–8] or functionalities [9,10]. A detailed review on existing SELEX methods has been published [11].
Figure 1.
Schematic of SELEX process which includes six major steps: library preparation, target incubation, target-binding strand separation, PCR amplification, single strand generation, and sequencing.
Aptamers have become widely used as recognition elements in biosensors for the detection of analytes such as ions, small molecules, and proteins [12,13]. They have many advantages relative to protein-based reagents such as antibodies, including ease of chemical synthesis with minimal batch-to-batch variation, low cost of production, high stability, amenability to chemical modification for sensing purposes, and tunable affinity and specificity [12,14]. Aptamer-based sensors for small molecules number in the hundreds [15], with targets including toxins [16], pharmaceuticals [17], illicit drugs [18,19], and endogenous compounds [20]. Many sensing strategies have been developed for aptamer-based sensors to achieve optical or electrochemical detection of targets [21–24]. These architectures are based on the interaction of aptamers that are labeled with chemical probes such as a fluorophore-quencher pair [24] or redox-active molecule [23] with specific analytes. The binding of the aptamer to the target results in a conformational change in the aptamer that is then transduced into a measurable signal due concomitant changes in environment of the probe. Aptamers with the ability to undergo target-induced conformational changes are generated by either destabilizing the aptamer through truncation or using a complementary sequence that partially blocks the binding domain.
Here, we describe previous and current advances in aptamer-based sensing, with a specific focus on the development of new detection modalities for small-molecule detection, and explore future directions for this field. First, we describe the use of enzymes in aptamer-based assays and aptamer engineering. We then review recent strategies for improving the sensitivity of aptamer-based sensors via cooperative binding. Finally, we discuss label-free approaches that enable simple, sensitive, and cost-effective detection of small molecules with fully-folded aptamers.
2. Exonuclease-assisted detection and engineering
With traditional aptamer-based assays, a single aptamer-target recognition event results in one signal. Although these methods are straightforward, they are generally not sensitive enough for real-world detection applications, in which targets may be present at very low concentrations. As such, several investigations have been performed in the use of nucleases to transduce one binding event into multiple signals (i.e. signal amplification) for achieving ultra-sensitive target detection.
One of the first works to utilize such enzymes in an aptamer-based assay employed nicking endonucleases: sequence-dependent nucleases that cleave the phosphodiester backbone of DNA at specific sites [25]. Willner et al. [26] used the nicking endonuclease Nt.BbvCI to develop an enzyme-based signal amplification strategy based on a cocaine-binding aptamer that possesses a partial Nt.BbvCI recognition site and an extended 5’-end polymerase template. In the absence of target, a short complementary DNA sequence hybridizes with the aptamer, partially blocking the target-binding domain. Aptamer-target binding leads to a large conformational change, displacing the complementary strand to form a duplexed 3’-end, such that polymerase extension can complete the recognition site. Nt.BbvCI cleavage of the replicated strand results in a nicked 3’-end, which is then used for a second round of polymerase extension to displace the nicked strand. This polymerization-nicking-strand-displacement cycle is continuously repeated, allowing for a single target to produce multiple displaced DNA strands that can then be quantified with fluorophore-quencher-modified molecular beacons. This strategy enabled detection of cocaine at concentrations as low as 5 μM within 60 minutes. Although this assay utilized enzyme amplification, its sensitivity was no more than 5-fold better than previous works [19,27]. This is probably due to that the use of strand displacement as an amplification initiator prohibits ultra-sensitive detection because the complementary DNA generally has higher affinity for the aptamer than the target itself, which results in strong competition between the two species for aptamer binding. In addition, the assay requires reporter-modified molecular beacon probes, which makes detection costly. Also, given that this enzyme is sequence-specific, designing aptamer constructs compatible with this enzyme can be challenging. There has since then been a concerted effort towards the development of enzyme-assisted assays which utilize sequence-independent enzymes.
DNase I is a sequence-independent endonuclease that can hydrolyze single- and double-stranded DNA [28]. However, due to its indiscriminate activity, DNase I digests aptamers regardless of their target-bound state and therefore cannot be directly employed for small-molecule detection. Advances in DNA-protective nanomaterials has allowed for the development of DNase I-based enzyme-assisted target recycling (EATR) assays. It has been shown that single-stranded DNA adsorbs strongly to graphene sheets through π-stacking interactions with the nucleobases [29]. Yang et al. [30] developed a DNase I-amplified strategy utilizing graphene and fluorescein-modified structure-switching aptamers that bind to ATP or cocaine. In the absence of target, the aptamers exist in a single-stranded state and readily adsorb onto the graphene surface, quenching the fluorophore and protecting them from DNase I digestion. Upon addition of the target, the aptamer folds into a duplexed structure and is released from the graphene surface, where it is digested by DNase I into mononucleotides with weak affinity for graphene, resulting in recovery of fluorescence [31]. The target is then released to bind to another graphene-adsorbed aptamer and continue this digestion cycle, thereby improving sensitivity. This assay achieved a calculated limit of detection (LOD) of 0.04 μM for ATP and 0.1 μM for cocaine after 2 hours of reaction, with signal gains that were five-fold higher than the same assay performed without DNase I-mediated amplification. This amplification strategy can also be used with other DNA protective nanomaterials such as carbon nanoparticles, single-walled carbon nanotubes, and magnetic graphene. DNase I has the advantage of performing well in a variety of physicochemical conditions. However, this strategy cannot be readily applied to aptamers that do not have structure-switching functionality. Moreover, DNA-protective nanomaterials typically have high batch-to-batch variation [32,33]. Recently, sequence-independent enzymes capable of recognizing designed small-molecule-induced conformational changes such as endonuclease IV [34], exonuclease III (Exo III) [35], DNazymes [36] have been incorporated into EATR methods for ultra-sensitive detection without the need for these nanomaterials.
In traditional EATR assays, target binding triggers digestion of the aptamer, resulting in a partially shortened sequence or complete digestion. Such methods require modification of aptamers with a signal reporter to quantify its target. To overcome this limitation, label-free exonuclease protection assays have also been developed. In these assays, target binding inhibits nuclease degradation of aptamers, whereas unbound aptamers are digested into mononucleotides. For example, Lou et al. [37] have utilized the enzyme exonuclease I (Exo I) in this assay format to detect a variety of analytes. Exo I digests single-stranded DNA in the 3’ to 5’ direction but has no activity on double-stranded DNA [38]. The authors engineered a structure-switching cocaine-binding aptamer that exists in a single-stranded state in the absence of cocaine and can be digested by Exo I into mononucleotides. In the presence of cocaine, the aptamer undergoes a large conformational change into a double-stranded structure, which resists digestion by Exo I. Following digestion, the remaining oligonucleotides can be easily quantified by commercially available DNA-binding dyes, such as SYBR Gold. This dye binds to intact oligonucleotides with high affinity and emits strong fluorescence when bound, but it is unable to bind mononucleotides [39]. This simple and label-free assay achieved a LOD of 5 μM cocaine within 40 minutes. Although exonuclease protection assays present an attractive label-free alternative to traditional EATR assays, they still require structure-switching aptamers as sensing elements, which typically have poor target-binding affinities, mitigating assay sensitivity.
We have recently found that inhibition of exonucleases by aptamer-target binding can also be utilized to engineer structure-switching aptamers. SELEX techniques usually use target binding as the sole selection force, and are thus ill-suited for specifically isolating the small subset of aptamers that undergo target-induced conformational changes [9,40]. Therefore, structure-switching aptamers are typically engineered after the selection process. This requires identification of the aptamer’s target-binding domain through the aid of programs such as mfold [41] and Nupack [42], or through enzymatic/chemical footprinting assays [43–47] followed by a multi-stage, expensive, trial-and-error process of sequence analysis, truncation, and chemical modification [24,48,49].
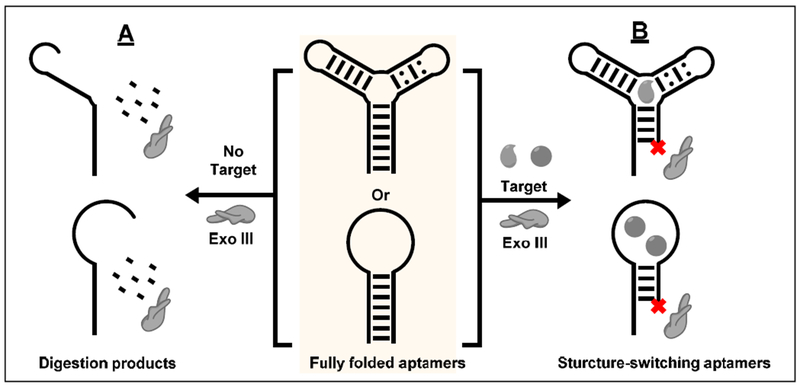
We developed a simple, single-step truncation strategy based on Exo III to rapidly generate structure-switching aptamers for use in folding-based aptamer sensors. Exo III is an exonuclease that digests double-stranded DNA from the 3’-to-5’ direction. Previous works have shown that Exo III digests target-bound aptamers but not unbound aptamers [50]. Contrary to this finding, we determined that Exo III digests aptamers into short single-stranded products in the absence of their target (Fig. 2A), but the binding of a small molecule to its respective aptamer halts digestion a few bases prior to the target-binding domain (Fig. 2B). The resulting truncated aptamers possess structure-switching functionality with similar binding affinities to their parent aptamers. We have demonstrated this concept with aptamers exhibiting a diverse range of secondary structures, binding to targets including adenosine triphosphate (ATP), cocaine, and dehydroisoandrosterone-3-sulfate (DIS). We have used the truncated ATP- and cocaine-binding aptamers to fabricate folding-based sensors for fluorescent and electrochemical detection of these targets with LODs of 2.5 μM and 1 μM, respectively [51]. Based on these findings, it is possible that other exonucleases can also be used to truncate aptamers. For example, it would be interesting to digest aptamers with both 3’-to-5’ and 5’-to-3’ exonucleases in order to derive fully minimized structure-switching aptamers for biosensing and home in more precisely on the approximate location of the target-binding domain by digesting from both termini.
Figure 2.
Scheme of Exo III-mediated aptamer truncation. (A) In the absence of target, Exo III digests aptamers into short oligonucleotides. (B) In the presence of target, Exo III digestion halts a few nucleotides prior to the target-binding domain, generating a structure-switching aptamer.
3. Improving sensitivity via cooperative target binding
Tuning the sensitivity of aptamer-based assays is crucial for real-world sensing applications. For example, sensors based on the well-known cocaine aptamer have detection limits in the micromolar range [18], which is unsuitable for the detection of this drug in biological samples, where the concentration is typically in the sub-micromolar regime. Numerous avenues have been explored to manipulate assay sensitivity and dynamic range, including aptamer affinity enhancement or attenuation via sequence engineering or rational mutation and signal amplification [52–55]. Our group has developed a strategy based on the concept of cooperative binding with aptamers. Cooperative binding of ligands is commonly observed with receptors in nature [56,57]. One often cited example is hemoglobin, which binds to molecular oxygen in an increasingly cooperative manner in high-oxygen environments [57]. Plaxco et al. [58] initially showed that cooperative binding can be introduced into nucleic acids by developing fluorophore-quencher-modified stem-loop DNA probes with two identical binding sites that are each complementary to the same target sequence. Initially, the fluorophore is near the quencher, resulting in no fluorescence. Binding of one target strand promotes partial disturbance of the stem, which allows binding to a second target strand with much higher affinity, opening the stem and resulting in a fluorescent signal. Using a ‘symmetric’ probe design that positions both binding sites adjacent to one another, the authors observed near-perfect cooperativity, with a Hill coefficient of 1.94. Plaxco et al. then extended this concept to aptamers [59]. Their approach involved engineering single-stranded constructs consisting of two aptameric domains linked in tandem, with a long single-stranded domain that entropically promoted disassembly of the whole construct. In the presence of the aptamer’s target, the aptameric domains assemble in a cooperative manner, with one target-binding event promoting binding by the second target (i.e. positive cooperativity). They demonstrated this concept with thymine-rich constructs that bind the mercury (II) cation, and with aptamers that bind doxorubicin and cocaine, and showed that this approach conferred control over the target-responsiveness of their aptamers. Nevertheless, engineering cooperativity via high entropy costs from disordered domains came at the price of dramatically reduced target affinity, which rendered the resulting constructs unsuitable for sensing purposes.
We have developed an alternative engineering approach [60] to introduce cooperativity into split aptamers, achieving levels of improved sensitivity that could be of potential benefit for sensing applications. Aptamers can be split from a single strand into two [24] or three [61] fragments. In the absence of target, the fragments are unable to assemble, but retain their capacity for target recognition and can successfully reassemble into a complex secondary structure in the presence of target [24,61]. Split-aptamer-based assays have certain advantages over other assay platforms, including low background signal due to the thermodynamic unfavourability of strand assembly in the absence of target. However, such assays usually have low sensitivity because split aptamers tend to have far lower binding affinity relative to the parent aptamer from which they were derived. This is actually an asset in the context of cooperative binding-based sensors. Cooperativity does not improve the sensitivity of single-stranded structure-switching aptamers because the aptamers need to be thoroughly destabilized to prevent background folding, which significantly decreases target affinity, However, since split aptamers are already destabilized, cooperativity makes it easier to achieve target-induced assembly without increasing background, thereby improving sensitivity.
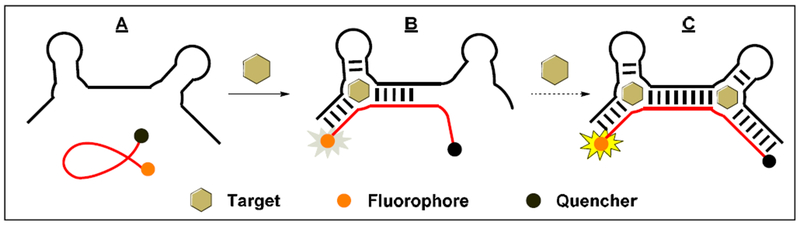
As a demonstration, we developed cooperative-binding split aptamers (CBSAs) from small-molecule-binding aptamers to achieve more sensitive detection [60,62,63]. We first engineered a cocaine-binding CBSA consisting of two target-binding domains linked in tandem by a 6 base-pair (bp) bridge with 5-bp terminal stems. One CBSA strand contains a terminal fluorophore-quencher pair that are in close proximity in the absence of target due to the flexibility of the strand, resulting in no fluorescence (Fig. 3A). The initial target-binding event induces assembly of the split aptamer (Fig. 3B), and facilitates subsequent target binding at the second binding domain, separating the fluorophore from the quencher and producing a fluorescent signal (Fig. 3C). Compared with split aptamers with a single target-binding domain, CBSAs exhibit higher target affinity and far more responsive target-induced aptamer assembly, enabling ultra-sensitive target detection. For example, with our cocaine-binding CBSA, we were able to detect cocaine with a LOD of 50 nM in 10% saliva within 15 minutes [60]. This could not be achieved with any conventional split-aptamer-based sensors [24,61,64]. Notably, the assay’s sensitivity makes it amenable for roadside detection of cocaine in saliva at recommended cutoff levels in the European Union [65].
Figure 3.
Cooperative-binding split aptamer (CBSA)-based assay for small-molecule detection. (A) In the absence of target, both CBSA fragments remain separate, and the fluorophore on the labeled strand is quenched due to close proximity to a quencher. (B) One target binding assembles both CBSA fragments which (C) promotes cooperative binding of a second target, separating fluorophore and quencher and thus producing an increase in fluorescence.
We then explored the possibility of improving assay sensitivity even further by utilizing the EATR strategy with CBSAs. To do so, we inserted an abasic site in the middle of one of the strands of the CBSA to enable cleavage by the apurinic/apyrimidinic endonuclease activity of Exo III [62]. The presence of target promotes aptamer assembly, enabling Exo III cleavage of the target-CBSA complex at the abasic site and recycling of the target, thereby amplifying the signal [62]. We demonstrated this approach by engineering a new CBSA for DIS. We achieved a LOD as low as 1 μM for DIS in 50% urine after 30 minutes, which is 100-fold more sensitive than could be achieved without EATR-mediated signal amplification. We have also used this strategy to achieve visual detection of cocaine by incorporating a CBSA into a gold nanoparticle-based colorimetric assay, which achieved a LOD of 10 μM, with naked-eye visualization within 20 minutes [62].
To perform label- and enzyme-free visual detection of small molecules with CBSAs, we recently developed ‘CBSAzymes’. These combine a CBSA module with a horseradish-peroxidase-mimicking G-quadruplex split DNAzyme module [63]. The presence of target promotes CBSA assembly, which in turn induces assembly of the split DNAzyme module; this enables hydrogen peroxide-mediated oxidation of colorless substrates such as 2,2’-azino-bis(3-ethylbenzthiazoline)-6-sulfonic acid (ABTS) into colored products for visual detection [63]. We demonstrated this approach with a cocaine-binding CBSAzyme, which allowed for label-free visual detection of 10 μM cocaine within 5 minutes. The generalizability of this strategy was shown by developing another CBSAzyme with an aptamer we recently isolated for methylenedioxypyrovalerone (MDPV), a designer drug from the synthetic cathinone family. In general, the development of assays for these drugs has been impeded by the lack of bioreceptors that can recognize members of this family [66]. By developing a CBSAzyme based on an aptamer that binds to MDPV, we were able to detect this drug as well as 10 other synthetic cathinones at micromolar levels in biological samples [63].
We are continuing to study cooperative-binding aptamers and their advantages as well as their applicability in other assay formats, such as electrochemical sensors [23], which offer a powerful strategy for achieving increased sensitivity for analyte detection in complex samples. To date, all engineered cooperative-binding aptamers have been derived from three-way-junction-structured aptamers, and there also remains a need for strategies to engineer similar cooperativity into aptamers with other common secondary structures such as stem-loops and G-quadruplexes.
4. Label-free sensing with fully folded aptamers
Recently, several strategies have been developed for rapid and sensitive analyte detection without the need for aptamers to undergo target-induced conformational changes. These typically employ a fully folded aptamer in combination with a signaling probe such as a dye molecule [67], exonuclease [68], or the target itself [69] to report target-binding events. This eliminates the need for laborious sequence engineering, and such assays have high sensitivity due to the preserved target affinity of fully folded aptamers relative to their truncated derivatives [49,52] or those employed in strand-displacement assays [70]. The need for specific signaling probes is the major drawback of these methods, but several new strategies have been developed to improve generality.
4.1. Aptamer-based dye-displacement assays
Dye-displacement assays rely on small-molecule dyes that can associate with the aptamer-binding domain [10,67,71–73]. The target displaces the dye from the binding domain, resulting in a change in absorbance or fluorescence that can be read instrumentally or observed with the naked eye. These assays are highly effective, as they are label-free, simple, and have rapid turnaround times and high sensitivity. This is because aptamers typically bind to the dye and target with similar affinities, and so target-induced dye-displacement is more thermodynamically and kinetically feasible than aptamer folding. As a result, dye-displacement assays can achieve a LOD that is 10-fold lower than the KD of the aptamer being used, and are usually more sensitive than assays using structure-switching aptamers [10,67,71–73].
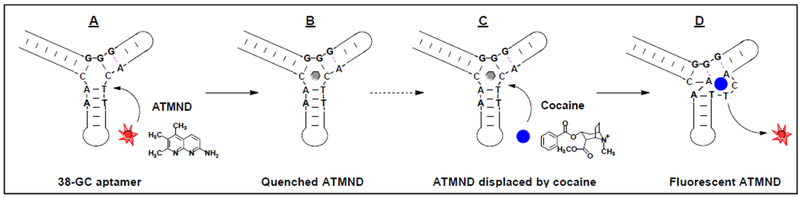
Stojanovic and Landry described the first aptamer-based dye-displacement assay in pioneering work from 2002, using a cocaine-binding aptamer and the cyanine dye diethylthiotricarbocyanine iodide (Cy7) [67]. The dye mainly exists as two forms - a monomer and a dimer - which have absorbance maxima of 760 and 670 nm, respectively. Cy7 monomers bind within the three-way-junction-structured binding domain of the aptamer, but can readily be displaced by cocaine, resulting in a decrease in the monomer peak within one minute. The sensitivity of this assay enabled cocaine detection at concentrations as low as 0.5 μM with 15-minute incubation. At higher aptamer and dye concentrations, the presence of 500 μM cocaine was able to induce formation of a visible dimer precipitate. However, visual detection times were quite long, requiring 12 hours for aggregation of the displaced dye. In other work, Wang et al. demonstrated a dye-displacement assay using an ATP aptamer and a ruthenium-based organometallic dye [74]. The dye binds to the G-quadruplex structured aptamer in the absence of target, which results in a large fluorescent signal. ATP displaces the dye from the aptamer, resulting in a concentration-dependent reduction in fluorescence. This method was sensitive, specific, and rapid, enabling detection of as low as 20 nM ATP in physiological buffer. More recently, we developed a highly sensitive dye-displacement assay utilizing 2-amino-5,6,7-trimethyl-1,8-naphthyridine (ATMND) as a signal reporter for sensitive cocaine detection [71]. The dye is weakly fluorescent when bound to the aptamer and is readily displaced by the target within seconds, where it fluoresces strongly in solution (Fig. 4). This assay enabled detection of cocaine at concentrations as low as 0.2 μM, which is 50-fold more sensitive than assays based on structure-switching aptamers [19].
Figure 4.
A dye-displacement assay using a cocaine-binding aptamer and ATMND for rapid detection of cocaine. Figure is reprinted from ref. [71] with permission, copyright 2014 American Chemical Society.
Although these assays have proven highly effective for individual targets, generalization remains a problem. In their original work, Stojanovic and Landry underscored the difficulty in finding dyes that were amenable for use in their assay: after screening at least 35 dyes, only Cy7 proved suitable [67]. Pei and Stojanovic provided further insights on the generality of this platform by investigating the fluorescent dye thiazole orange with a variety of DNA and RNA aptamers with various secondary structures that bind to small molecule and protein targets [75]. Thiazole orange bound to several aptamers to form a fluorescent complex, with displacement of the dye by the target resulting in a reduction in fluorescence. Sensitivities for these successful candidates were in the nanomolar to sub-micromolar range, with turnaround times of 40 minutes. However, they found that this dye was compatible with only eight of 19 tested aptamers in this assay— again, highlighting the difficulty in finding generally applicable dyes for displacement assays.
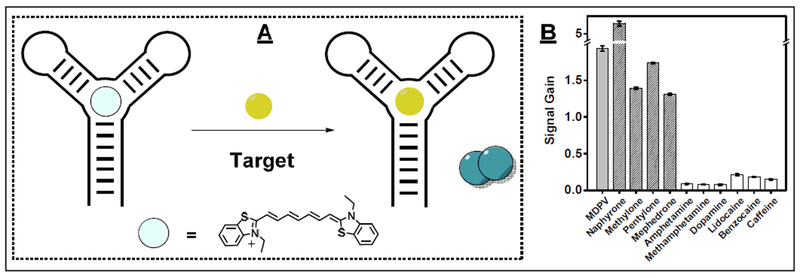
We recently investigated whether it was possible to generalize the platform by using Cy7. We systematically determined that Cy7 monomers can generally bind within the target-binding domains of three-way-junction-structured aptamers, and that target binding can in turn displace the dye to induce dimer formation and a ratiometric change in absorbance amenable for analyte detection (Fig. 5A). We isolated a three-way-junction-structured aptamer that binds to MDPV and found that it could readily be used in a dye-displacement format with Cy7 without any need for sequence engineering. This enabled detection of 300 nM MDPV as well as four other synthetic cathinones (pentylone, naphyrone, mephedrone, and methylone) within seconds (Fig. 5B) [10]. However, the aptamer was also found to bind interferents and unrelated molecules such as cocaine and DIS [10]. This coincides with previous reports showing that three-way-junction aptamers have poor specificity [76,77].
Figure 5.
A dye-displacement assay using an MDPV-binding aptamer and Cy7 for the detection of MDPV. (A) Scheme for this assay and (B) assay response to MDPV, four other synthetic cathinones (hatched bars), and six interferents (white bars). Panel B reprinted from ref. [10] with permission, copyright 2018 Oxford University Press.
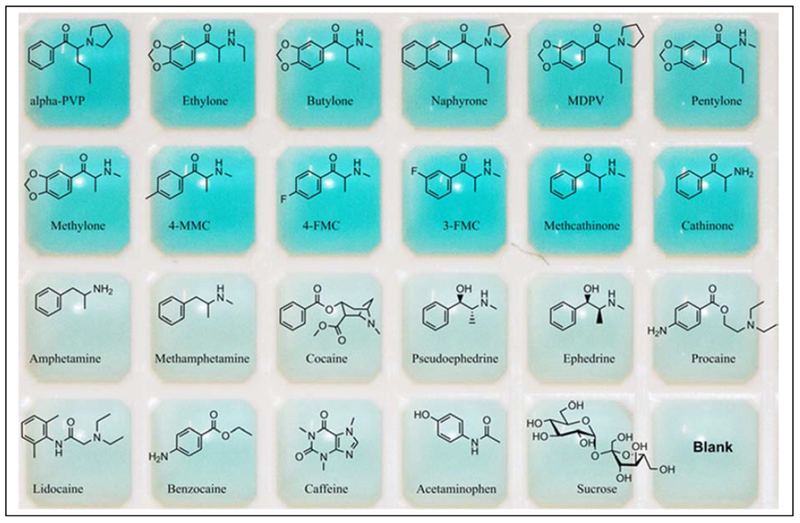
We next explored whether Cy7 is compatible with other aptamers with secondary structures other than three-way junctions. In our recent work, we further expanded the generality of the Cy7-displacement assay by demonstrating its applicability with stem-loop structured aptamers [7]. We isolated a cross-reactive hairpin-structured aptamer binding to synthetic cathinones [7], and showed that it could bind to Cy7 such that targets can displace the dye from the target-binding domain of the aptamer to produce an instantaneous reduction of monomer absorbance and increase in dimer absorbance. With this assay, we were able to detect low nanomolar concentrations of 12 different synthetic cathinones, even in biological samples. Impressively, the assay yielded no response to 11 interferents including other drugs of abuse and prescription medications, with some bearing considerable structural similarity to true targets. By fine-tuning dye and aptamer concentrations, we were able to obtain a clear-to-blue color change that can be observed with the naked eye upon the addition of target within seconds, with a LOD of ~6 μM (Fig. 6).
Figure 6.
Photograph of aptamer-based Cy7-displacement assay for 12 synthetic cathinones (top two rows) and 11 interferents and a negative control (bottom two rows), taken a few seconds after addition of ligand.
Much work is still required to make dye-displacement assays truly generalizable. We are currently exploring whether Cy7 is generally compatible with aptamers with other higher order structures such as G-quadruplexes. We are also trying to find or synthesize other dyes that can generally bind to aptamers with specific secondary structures to enable straightforward assay development with structured libraries. If these efforts prove successful, we envision dye-displacement assays becoming an assay of choice for instrument-free aptamer-based optical detection.
4.2. Exonuclease-mediated aptamer-based assay
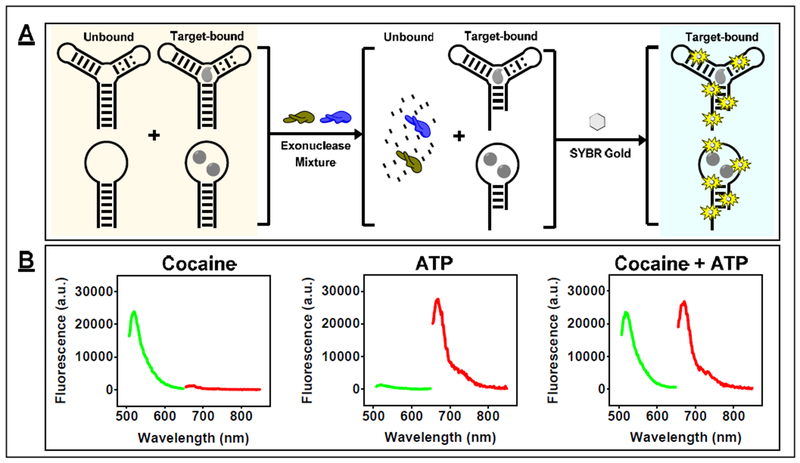
Since the dye-displacement assay is not compatible with all aptamers, we have also developed an enzyme-based method that is generally applicable for aptamers with varying sequences and secondary structures. This approach uses a mixture of Exo III and Exo I with fully folded high-affinity aptamers to achieve ultra-sensitive small-molecule detection [68]. In the absence of target, Exo III and Exo I digest double- and single-stranded regions of DNA aptamers, respectively, allowing for complete digestion of aptamers with hairpin and three-way-junction structures into mononucleotides. Target binding to the aptamer inhibits digestion by both enzymes a few nucleotides prior the target-binding domain of the aptamer, resulting in partially-digested oligonucleotide products that can subsequently be quantified with SYBR Gold (Fig. 7A). We used this assay to detect a variety of small molecules such as DIS, cocaine, and ATP, achieving LODs of 0.5, 0.1, and 0.25 μM, respectively, within 15–20 minutes. Notably, this assay works equally well in biological samples, enabling detection of DIS and cocaine in 50% urine and 10% saliva, respectively, with the same LOD as in buffer.
Figure 7.
Label-free small-molecule detection with Exo III and Exo I. This assay is compatible with multiple readouts. (A) For example, SYBR Gold can be used for generic target detection. (B) Alternatively, molecular beacons can be used as signal reporters to detect multiple targets simultaneously in a mixture. Panel B reprinted from ref. [68] with permission, copyright 2018 American Chemical Society.
Importantly, the final oligonucleotide quantification step can be tailored for a desired detection goal. For example, since the sequence of the inhibition products for each target were known, we were able to utilize fluorophore-quencher-modified molecular beacons to quantify both cocaine and ATP in a mixture simultaneously (Fig. 7B). In the future, it should become feasible to incorporate other advanced DNA detection methods and strategies into this assay, such as microarrays or DNA amplification assays, for specific analytical applications.
4.3. Aptamer field-effect transistor (FET) sensors
Receptor-modified FETs have recently gained attention in the biosensing field. The conductivity of the semiconductor channel in FETs is extremely sensitive to changes in the local electric field. When the surface of a FET is modified with a specific bioreceptor such as an aptamer, binding of charged target molecules at the surface is sufficient to generate an electrical signal [78]. FET-based sensors can achieve ultra-high sensitivity, partially because of the nonlinear behavior of the semiconductor channel. Since the charge on the targets is being employed for signal transduction, high-affinity fully folded aptamers can be directly employed in the sensor. Moreover, FETs offer a broad dynamic range spanning several orders of magnitude, which cannot be achieved with most sensing platforms [79]. For example, Weiss et al. demonstrated detection of positively-charged dopamine using an aptamer-modified FET with a LOD of 10 pM and a dynamic range from 10 pM to 1 μM [80]. Aptamer-modified FETs have some significant limitations, however. First, uncharged targets have minimal effect on the semiconductor, and thus cannot be effectively measured [81]. Second, semiconductor transconductance is only sensitive to variations in charge density within the electrical double layer, but the thickness of this layer dramatically decreases as solution ionic strength increases. This thickness drops below 1 nm in physiological fluids, which makes target detection challenging in these conditions [82].
Recent work has shown the possibility of remedying both limitations, based on the determination that small-molecule binding can introduce a ternary structure change in fully-folded aptamers modified onto the FET surface. This rearrangement of the negatively-charged aptamer phosphodiester backbone near the semiconductor surface can then be detected by FET even if the binding event itself occurs outside the electrical double layer. Andrews et al. have developed a FET-based aptamer sensor for the detection of both charged and uncharged small molecules [83]. They first isolated aptamers that bind to various endogenous compounds, including dopamine, serotonin, glucose, and sphingosine-1-phosphate. These aptamers were then covalently attached to the surface of an indium-oxide semiconductor via silane chemistry. The binding of the target to an aptamer induces a subtle conformational change that either orients the aptamer closer to or further away from the surface, which decreases or increases the transconductance of the sensor, respectively. These sensors enabled detection of the aforementioned analytes at femtomolar levels in buffer and picomolar levels in biological samples. The generalizability of this strategy for other small-molecule-binding aptamers remains to be assessed, however. This will not be easy, due to the need for specialized equipment and techniques for fabrication of aptamer-modified FETs, which are not accessible for most labs.
5. Future Directions
Aptamers have many attractive features that make them suitable for the creation of sensitive, specific, easy-to-use biosensors. This has made many speculate aptamers to be up-and-coming replacements of traditional protein bioreceptors like antibodies. However, this has yet to occur. Here, we have described several new developments in different aptamer sensors platforms with enhanced sensitivities and excellent performance in biomatrices that are focused on bringing the field towards this end (Table 1). However, we posit that advances on other fronts are needed prior to aptamers becoming mainstream. These matters are related to improvement of the binding affinity of aptamers and the design of practical, more sensitive and simple sensors that can function outside of laboratory settings.
Table 1.
Various aptamer-based sensing platform described in this work.
| Detection Strategy | Sensing Element | Target | LOD | Detection time | Ref |
|---|---|---|---|---|---|
| Enzyme-assisted signal amplification | Aptamer + cDNA | Cocaine | 5 μM | 70 min | 26 |
| Enzyme-assisted target recycling | Structure-switching aptamer | ATP | 0.1 μM | 120 min | 30 |
| Structure-switching aptamer | Cocaine | 0.1 μM | 120 min | 30 | |
| Split aptamers | Adenosine | 5 pM | 2.5 h | 34 | |
| Split aptamers | Cocaine | 10 pM | 2.5 h | 34 | |
| Structure-switching aptamer | ATP | 0.25 μM | 15 min | 35 | |
| Structure-switching aptamer | Cocaine | 50 nM | 15 min | 35 | |
| Strand displacement | Aptamer + cDNA | Adenosine | 100 μM | 6 min | 27 |
| Aptamer + cDNA | Cocaine | 50 μM | 5 min | 27 | |
| Split-aptamer-based | Split aptamers | Cocaine | 10 μM | 60 min | 24 |
| Split aptamers | ATP | 10 μM | 60 min | 24 | |
| Split aptamers | Cocaine | 2 μM | 18 min | 61 | |
| Split aptamers | ATP | 0.1 μM | 7 min | 61 | |
| Split aptamers | Cocaine | 20 μM | 5 min | 64 | |
| Cooperative-binding split aptamer | Cocaine | 50 nM | 15 min | 60 | |
| Split-aptamer-based enzyme-assisted target recycling | Cooperative-binding split aptamer | DIS | 0.5 μM | 30 min | 62 |
| Cooperative-binding split aptamer | Cocaine | 0.2 μM | 30 min | 62 | |
| Split aptamer and DNAzyme-based signal amplification | Cooperative-binding split aptamer | MDPV | 3 μM | 15 min | 63 |
| Cooperative-binding split aptamer | Cocaine | 1 μM | 15 min | 63 | |
| Dye displacement | Fully-folded aptamer | Cocaine | 0.5 μM | 15 min | 67 |
| Fully-folded aptamer | Cocaine | 0.2 μM | Seconds | 71 | |
| Fully-folded aptamer | ATP | 20 nM | N/A | 74 | |
| Fully-folded aptamer | AMP | 9.8 nM | 40 min | 75 | |
| Fully-folded aptamer | GTP | 0.39 μM | 40 min | 75 | |
| Fully-folded aptamer | MDPV | 0.3 μM | Seconds | 10 | |
| Fully-folded aptamer | MDPV | 0.25 μM | Seconds | 7 | |
| Exonuclease inhibition | Structure-switching aptamer | Cocaine | 5 μM | 40 min | 37 |
| Fully-folded aptamer | Cocaine | 1 μM | 15 min | 51 | |
| Fully-folded aptamer | DIS | 0.5 μM | 15 min | 68 | |
| Fully-folded aptamer | ATP | 0.25 μM | 20 min | 68 | |
| Fully-folded aptamer | Cocaine | 0.1 μM | 25 min | 68 | |
| Aptamer-based field-effect transistor | Fully-folded aptamer | Dopamine | 10 pM | Seconds | 80 |
| Fully-folded aptamer | Dopamine | 10 fM | Seconds | 83 | |
| Fully-folded aptamer | Serotonin | 10 fM | Seconds | 83 | |
| Fully-folded aptamer | Glucose | 10 fM | Seconds | 83 | |
| Fully-folded aptamer | Sphingosine-1-phosphate | 10 fM | Seconds | 83 |
While the affinity of aptamers for high molecular weight targets such as proteins is sufficient for the detection of these analytes, this is not case for aptamers targeting small molecules. There are some solutions to this problem. For one, the development of new SELEX methods specifically tailored for isolating small-molecule-binding aptamers with high affinities is in need. Aptamers are typically isolated through methods whereby the target or library is immobilized on surfaces such as agarose or magnetic beads. This can limit the potential interaction of the target/library strand with their binding partner due to a mixture of conformational restraints, steric hinderance, or functional group masking. The outcome of this is aptamers with low affinity for their small-molecule target. Therefore, the development of a ‘bead-free’ SELEX method that allows the library and target to interact freely in solution would be of great benefit. One example is the use of graphene oxide as a partitioning agent to isolate aptamers without the need for immobilization of target or library [84,85]. Another solution that has recently become popular is the use of high-throughput sequencing to identify enrichment of high affinity binders throughout selection rounds [86]. Also, modified nucleotides with novel functional groups not found in nucleic acids have been incorporated in libraries or isolated aptamers as a means of increasing structural diversity and opportunities for ligand binding to improve aptamer-target interactions. This has proven successful in certain cases; however, the use of modified nucleotides remains challenging and it is easier to work with and synthesize aptamers containing natural nucleotides [87].
In addition, there must be a movement away from prototype aptamer-based sensors and a push towards developing sensors that can be operated simply and realistically for use on-site or at the point-of-care. Such sensors should require as few reagents and steps (ideally, one single-step detection) as possible, with minimal or no instrumental requirements and rapid turnaround times. Some of the assay formats we have presented herein have many of these characteristics, such as the CBSA-based and dye-displacement assays. However, many reported aptasensors stray from this ideal paradigm, requiring multiple steps, special and sensitive reagents, and, in cases where instrumentation is required, no propositions or suggestions for simple and cost-effective equipment. For example, many E-AB sensors have been developed for the detection of a variety of compounds, wherein sensor performance is demonstrated using benchtop electrochemistry stations and gold disc electrodes or complicated microfluidic operating systems. This setup cannot be readily adapted for on-site applications. A solution to this would entail the development of new paper-based E-AB compatible with portable handheld potentiostats. This paper-based technology should be adapted for colorimetric aptamer-based assays.
6. Conclusion
As aptamers reach their thirtieth anniversary[1,2], the field continues to grow. Though the number of aptamer-based sensors continuously rises, the development of aptamer isolation and characterization techniques, as well as new engineering and sensing strategies, lags. This is particularly true for small-molecule detection, where challenges remain for sensitive, specific, and facile detection in real-world analytical contexts. Here, we have described new methodologies for engineering aptamer-based sensors for small-molecule targets. These include methods for signal amplification and introducing structure-switching functionality for folding-based sensors using enzymes, the use of cooperativity to develop ultra-sensitive assays for small-molecule detection, and label-free assays that utilize fully folded high-affinity aptamers. We envision that continuing progress in refining these powerful technologies, advances in aptamer isolation and characterization, along with progress in related fields such as nanotechnology, biotechnology, and synthetic biology, will enable the field to overcome roadblocks encumbering the practical and routine use of small-molecule-binding aptamers for molecular detection.
Highlights.
Enzyme-based signal amplification and engineering for aptamers.
Improving the sensitivity of small-molecule detection with cooperative binding aptamers.
Rapid and label-free dye-displacement aptamer-based assays.
Generalizable aptamer-based small-molecule detection via exonuclease digestion.
Acknowledgements
Work in the Xiao lab was supported by the National Institutes of Health - National Institute on Drug Abuse [R15DA036821] and the National Institute of Justice [2013-DN-BX-K032, 2015-R2-CX-0034, and 2016-DN-BX0167X]. O.A. acknowledges support from the Presidential Fellowship awarded by the University Graduate School of Florida International University.
Abbreviations
- SELEX
systematic evolution of ligands by exponential enrichment
- EATR
enzyme-assisted target recycling
- CBSA
cooperative-binding split aptamer
- Exo I
exonuclease I
- Exo III
exonuclease III
- ATP
adenosine triphosphate
- DIS
dehydroisoandrosterone-3-sulfate
- MDPV
3,4-methylenedioxypyrovalerone
- LOD
limit of detection
- Cy7
diethylthiotricarbocyanine iodide
- ATMND
2-amino-5,6,7-trimethyl-1,8-naphthyridine
- ABTS
2,2’-azino-bis(3-ethylbenzthiazoline)-6-sulfonic acid
- FET
field-effect transistor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Ellington AD, Szostak JW, In vitro selection of RNA molecules that bind specific ligands., Nature. 346 (1990) 818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- [2].Tuerk C, Gold L, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase., Science. 249 (1990) 505–510.doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- [3].Yang K-A, Pei R, Stojanovic MN, In vitro selection and amplification protocols for isolation of aptameric sensors for small molecules, Methods. 106 (2016) 58–65. doi: 10.1016/j.ymeth.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sanger F, Nicklen S, Coulson AR, DNA sequencing with chain-terminating inhibitors, Proc. Natl. Acad. Sci. U. S. A 74 (1977) 5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cho M, Xiao Y, Nie J, Stewart R, Csordas AT, Oh SS, Thomson JA, Soh HT, Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].White R, Rusconi C, Scardino E, Wolberg A, Lawson J, Hoffman M, Sullenger B, Generation of species cross-reactive aptamers using “Toggle” SELEX, Mol. Ther 4 (2001) 567–573. doi: 10.1006/mthe.2001.0495. [DOI] [PubMed] [Google Scholar]
- [7].Yang W, Yu H, Alkhamis O, Liu Y, Canoura J, Fu F, Xiao Y, In vitro isolation of class-specific oligonucleotide-based small-molecule receptors, Nucleic Acids Res. 47 (2019) e71. doi: 10.1093/nar/gkz224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jenison RD, Gill SC, Pardi A, Polisky B, High-resolution molecular discrimination by RNA, Science. 263 (1994) 1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- [9].Nutiu R, Li Y, In vitro selection of structure-switching signaling aptamers, Angew. Chem., Int. Ed 44 (2005) 1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- [10].Yu H, Yang W, Alkhamis O, Canoura J, Yang K-A, Xiao Y, In vitro isolation of small-molecule-binding aptamers with intrinsic dye-displacement functionality, Nucleic Acids Res. 46 (2018) e43. doi: 10.1093/nar/gky026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mascini M, Palchetti I, Tombelli S, Nucleic acid and peptide Aptamers: fundamentals and bioanalytial aspects, Angew. Chem., Int. Ed 51 (2012) 1316–1332. doi: 10.1002/anie.20100660. [DOI] [PubMed] [Google Scholar]
- [12].Dunn MR, Jimenez RM, Chaput JC, Analysis of aptamer discovery and technology, Nat. Rev. Chem 1 (2017) 0076. doi: 10.1038/s41570-017-0076. [DOI] [Google Scholar]
- [13].Jayasena SD, Aptamers: An emerging class of molecules that rival antibodies in diagnostics, Clin. Chem 45 (1999) 1628–1650. doi: http://clinchem.aaccjnls.org/content/45/9/1628. [PubMed] [Google Scholar]
- [14].Toh SY, Citartan M, Gopinath SCB, Tang T-H, Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay, Biosens. Bioelectron 64 (2015) 392–403. doi: 10.1016/j.bios.2014.09.026. [DOI] [PubMed] [Google Scholar]
- [15].Song S, Wang L, Li J, Fan C, Zhao J, Aptamer-based biosensors, TrAC, Trends Anal. Chem. 27 (2008) 108–117. doi: 10.1016/j.trac.2007.12.004. [DOI] [Google Scholar]
- [16].Li X, Cheng R, Shi H, Tang B, Xiao H, Zhao G, A simple highly sensitive and selective aptamer-based colorimetric sensor for environmental toxins microcystin-LR in water samples, J. Hazard. Mater 304 (2016) 474–480. doi: 10.1016/j.jhazmat.2015.11.016. [DOI] [PubMed] [Google Scholar]
- [17].Tombelli S, Mascini M, Aptamers biosensors for pharmaceutical compounds, Comb. Chem. High Throughput Screen. 13 (2010) 641–649. doi: 10.2174/1386207311004070641. [DOI] [PubMed] [Google Scholar]
- [18].Stojanovic MN, de Prada P, Landry DW, Aptamer-based folding fluorescent sensor for cocaine, J. Am. Chem. Soc 123 (2001) 4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- [19].Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW, An electronic, aptamer-based small-molecule sensor for the rapid, label-free detection of cocaine in adulterated samples and biological fluids., J. Am. Chem. Soc 128 (2006) 3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- [20].Farjami E, Campos R, Nielsen JS, V Gothelf K, Kjems J, Ferapontova EE, RNA aptamer-based electrochemical biosensor for selective and label-free analysis of dopamine, Anal. Chem 85 (2013) 121–128. doi: 10.1021/ac302134s. [DOI] [PubMed] [Google Scholar]
- [21].Lee J-O, So H-M, Jeon E-K, Chang H, Won K, Kim YH, Aptamers as molecular recognition elements for electrical nanobiosensors, Anal. Bioanal. Chem 390 (2008) 1023–1032. doi: 10.1007/s00216-007-1643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Willner I, Zayats M, Electronic aptamer-based sensors, Angew. Chem., Int. Ed 46 (2007) 6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- [23].Xiao Y, Lubin AA, Heeger AJ, Plaxco KW, Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor, Angew. Chem., Int. Ed 44 (2005) 5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- [24].Stojanovic MN, de Prada P, Landry DW, Fluorescent sensors based on aptamer self-assembly, J. Am. Chem. Soc 122 (2000) 11547–11548. doi: 10.1021/ja0022223. [DOI] [PubMed] [Google Scholar]
- [25].Heiter DF, Lunnen KD, Wilson GG, Site-specific DNA-nicking mutants of the heterodimeric restriction endonuclease R.BbvCI, J. Mol. Biol 348 (2005) 631–640. doi: 10.1016/j.jmb.2005.02.034. [DOI] [PubMed] [Google Scholar]
- [26].Shlyahovsky B, Li D, Weizmann Y, Nowarski R, Kotler M, Willner I, Spotlighting of cocaine by an autonomous aptamer-based machine, J. Am. Chem. Soc 129 (2007) 3814–3815. doi: 10.1021/ja069291n. [DOI] [PubMed] [Google Scholar]
- [27].Liu J, Lu Y, Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles, Angew. Chem., Int. Ed 45 (2006) 90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- [28].Kunitz M, Crystalline deoxyribonuclease, J. Gen. Physiol 33 (1950) 349–362. doi: 10.1085/jgp.33.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Patil AJ, Vickery JL, Scott TB, Mann S, Aqueous stabilization and self-assembly of graphene sheets into layered bio-nanocomposites using DNA, Adv. Mater 21 (2009) 3159–3164. doi: 10.1002/adma.200803633. [DOI] [Google Scholar]
- [30].Lu C-H, Li J, Lin M-H, Wang Y-W, Yang H-H, Chen X, Chen G-N, Amplified aptamer-based assay through catalytic recycling of the analyte, Angew. Chem., Int. Ed 49 (2010) 8454–8457. doi: 10.1002/anie.201002822. [DOI] [PubMed] [Google Scholar]
- [31].Varghese N, Mogera U, Govindaraj A, Das A, Maiti PK, Sood AK, Rao CNR, Binding of DNA nucleobases and nucleosides with graphene, ChemPhysChem. 10 (2009) 206–210. doi: 10.1002/cphc.200800459. [DOI] [PubMed] [Google Scholar]
- [32].Zhong YL, Tian Z, Simon GP, Li D, Scalable production of graphene via wet chemistry: progress and challenges, Mater. Today. 18 (2015) 73–78. doi: 10.1016/j.mattod.2014.08.019. [DOI] [Google Scholar]
- [33].Zhu BY, Murali S, Cai W, Li X, Suk JW, Potts JR, Ruoff RS, Graphene and graphene oxide: synthesis, properties, and applications, Adv. Mater 22 (2010) 3906–3924. doi: 10.1002/adma.201001068. [DOI] [PubMed] [Google Scholar]
- [34].Li Q, Wang Y-D, Shen G-L, Tang H, Yu R-Q, Jiang J-H, Split aptamer mediated endonuclease amplification for small-molecule detection, Chem. Commun 51 (2015) 4196–4199. doi: 10.1039/C5CC00390C. [DOI] [PubMed] [Google Scholar]
- [35].Liu X, Freeman R, Willner I, Amplified fluorescence aptamer-based sensors using exonuclease III for the regeneration of the analyte, Chem. - Eur. J 18 (2012) 2207–2211. doi: 10.1002/chem.201103342. [DOI] [PubMed] [Google Scholar]
- [36].Liu J, Lu Y, A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles, J. Am. Chem. Soc 125 (2003) 6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- [37].Zheng D, Zou R, Lou X, Label-free fluorescent detection of ions, proteins, and small molecules using structure-switching aptamers, SYBR gold, and exonuclease I, Anal. Chem 84 (2012) 3554–3560. doi: 10.1021/ac300690r. [DOI] [PubMed] [Google Scholar]
- [38].Lehman IR, Nussbaum AL, The Deoxyribonucleases of Escherichia coli: On the Specificity of Exonuclease I (Phosphodiesterase), J. Biol. Chem 239 (1964) 2628–2636. [PubMed] [Google Scholar]
- [39].Tuma RS, Beaudet MP, Jin X, Jones LJ, Cheung C-Y, Yue S, Singer VL, Characterization of SYBR gold nucleic acid gel stain: A dye optimized for use with 300-nm ultraviolet transilluminators, Anal. Biochem 268 (1999) 278–288. doi: 10.1006/abio.1998.3067. [DOI] [PubMed] [Google Scholar]
- [40].McKeague M, DeRosa MC, Challenges and opportunities for small molecule aptamer development, J. Nucleic Acids. 2012 (2012) 748913. doi: 10.1155/2012/748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zuker M, Mfold web server for nucleic acid folding and hybridization prediction, Nucleic Acids Res. 31 (2003) 3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA, NUPACK: Analysis and design of nucleic acid systems, J. Comput. Chem 32 (2011) 170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- [43].Higuchi R, Krummel B, Saiki R, A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions, Nucleic Acids Res. 16 (1988) 7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brenowitz M, Senear DF, Kingston RE, DNase I footprint analysis of protein-DNA binding, Curr. Protoc. Mol. Biol 7 (1989) 12.4.1–12.4.16. doi: 10.1002/0471142727.mb1204s07. [DOI] [PubMed] [Google Scholar]
- [45].Christy B, Nathans D, DNA binding site of the growth factor-inducible protein Zif268, Proc. Natl. Acad. Sci. U. S. A 86 (1989) 8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fischer NO, Tok JB-H, Tarasow TM, Massively parallel interrogation of aptamer sequence, structure and function, PLoS One. 3 (2008) e2720. doi: 10.1371/journal.pone.0002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gao S, Zheng X, Jiao B, Wang L, Post-SELEX optimization of aptamers, Anal. Bioanal. Chem 408 (2016) 4567–4573. doi: 10.1007/s00216-016-9556-2. [DOI] [PubMed] [Google Scholar]
- [48].White RJ, Rowe AA, Plaxco KW, Re-engineering aptamers to support reagentless, self-reporting electrochemical sensors, Analyst. 135 (2010) 589–594. doi: 10.1039/b921253a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Neves MAD, Reinstein O, Johnson PE, Defining a stem length-dependent binding mechanism for the cocaine-binding aptamer. A combined NMR and calorimetry study, Biochemistry. 49 (2010) 8478–8487. doi: 10.1021/bi100952k. [DOI] [PubMed] [Google Scholar]
- [50].Rogers SG, Weiss B, Exonuclease III of Escherichia coli K-12, an AP endonuclease, Methods Enzym. 65 (1980) 201–211. doi: 10.1016/S0076-6879(80)65028-9. [DOI] [PubMed] [Google Scholar]
- [51].Wang Z, Yu H, Canoura J, Liu Y, Alkhamis O, Fu F, Xiao Y, Introducing structure-switching functionality into small-molecule-binding aptamers via nuclease-directed truncation, Nucleic Acids Res. 46 (2018) e81. doi: 10.1093/nar/gky305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Neves MAD, Reinstein O, Saad M, Johnson PE, Defining the secondary structural requirements of a cocaine-binding aptamer by a thermodynamic and mutation study, Biophys. Chem 153 (2010) 9–16. doi: 10.1016/j.bpc.2010.09.009. [DOI] [PubMed] [Google Scholar]
- [53].Kang D, Vallee-Belisle A, Porchetta A, Plaxco KW, Ricci F, Re-engineering electrochemical biosensors to narrow or extend their useful dynamic range, Angew. Chem., Int. Ed 51 (2012) 6717–6721. doi: 10.1002/anie.201202204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wu Z-S, Zhang S, Zhou H, Shen G-L, Yu R, Universal aptameric system for highly sensitive detection of protein based on structure-switching-triggered rolling circle amplification, Anal. Chem 82 (2010) 2221–2227. doi: 10.1021/ac901794w. [DOI] [PubMed] [Google Scholar]
- [55].Nonaka Y, Yoshida W, Abe K, Ferri S, Schulze H, Bachmann TT, Ikebukuro K, Affinity improvement of a VEGF aptamer by in silico maturation for a sensitive VEGF-detection system, Anal. Chem 85 (2013) 1132–1137. doi: 10.1021/ac303023d. [DOI] [PubMed] [Google Scholar]
- [56].Meyer T, Holowka D, Stryer L, Highly cooperative opening of calcium channels by inositol 1,4,5-trisphosphate, Science. 240 (1988) 653–656. doi: 10.1126/science.2452482. [DOI] [PubMed] [Google Scholar]
- [57].Bellelli A, Hemoglobin and cooperativity: Experiments and theories, Curr. Protein Pept. Sci 11 (2010) 2–36. doi: 10.2174/138920310790274653. [DOI] [PubMed] [Google Scholar]
- [58].Simon AJ, Vallee-Belisle A, Ricci F, Watkins HM, Plaxco KW, Using the population-shift mechanism to rationally introduce “Hill-type” cooperativity into a normally non-cooperative receptor, Angew. Chem., Int. Ed 53 (2014) 9471–9475. doi: 10.1002/anie.201403777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Simon AJ, Vallee-Belisle A, Ricci F, Plaxco KW, Intrinsic disorder as a generalizable strategy for the rational design of highly responsive, allosterically cooperative receptors, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 15048–15053. doi: 10.1073/pnas.1410796111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yu H, Canoura J, Guntupalli B, Lou X, Xiao Y, A cooperative-binding split aptamer assay for rapid specific and ultra-sensitive fluorescence detection of cocaine in saliva, Chem. Sci 8 (2017) 131–141. doi: 10.1039/C6SC01833E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zou R, Lou X, Ou H, Zhang Y, Wang W, Yuan M, Guan M, Luo Z, Liu Y, Highly specific triple-fragment aptamer for optical detection of cocaine, RSC Adv. 2 (2012) 4636–4638. doi: 10.1039/C2RA20307C. [DOI] [Google Scholar]
- [62].Yu H, Canoura J, Guntupalli B, Alkhamis O, Xiao Y, Sensitive detection of small-molecule targets using cooperative binding split aptamers and enzyme-assisted target recycling, Anal. Chem 90 (2018) 1748–1758. doi: 10.1021/acs.analchem.7b03625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Luo Y, Yu H, Alkhamis O, Liu Y, Lou X, Yu B, Xiao Y, Label-free, visual detection of small molecules using highly target-responsive multimodule split aptamer constructs, Anal. Chem 91 (2019) 7199–7207. doi: 10.1021/acs.analchem.9b00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhang J, Wang L, Pan D, Song S, Boey FYC, Zhang H, Fan C, Visual cocaine detection with gold nanoparticles and rationally engineered aptamer structures, Small. 4 (2008) 1196–1200. doi: 10.1002/smll.200800057. [DOI] [PubMed] [Google Scholar]
- [65].Gjerde H, Langel K, Favretto D, Verstraete AG, Estimation of equivalent cutoff thresholds in blood and oral fluid for drug prevalence studies, J. Anal. Toxicol 38 (2014) 92–98. doi: 10.1093/jat/bkt122. [DOI] [PubMed] [Google Scholar]
- [66].Namera A, Kawamura M, Nakamoto A, Saito T, Nagao M, Comprehensive review of the detection methods for synthetic cannabinoids and cathinones, Forensic Toxicol. 33 (2015) 175–194. doi: 10.1007/s11419-015-0270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Stojanovic MN, Landry DW, Aptamer-based colorimetric probe for cocaine, J. Am. Chem. Soc 124 (2002) 9678–9679. doi: 10.1021/ja0259483. [DOI] [PubMed] [Google Scholar]
- [68].Canoura J, Wang Z, Yu H, Alkhamis O, Fu F, Xiao Y, No structure-switching required: A generalizable exonuclease-mediated aptamer-based assay for small-molecule detection, J. Am. Chem. Soc 140 (2018) 9961–9971. doi: 10.1021/jacs.8b04975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Shoara AA, Slavkovic S, Donaldson LW, Johnson PE, Analysis of the interaction between the cocaine-binding aptamer and its ligands using fluorescence spectroscopy, Can. J. Chem 95 (2017) 1253–1260. doi: 10.1139/cjc-2017-0380. [DOI] [Google Scholar]
- [70].Nutiu R, Li Y, Structure-switching signaling aptamers, J. Am. Chem. Soc 125 (2003) 4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- [71].Roncancio D, Yu H, Xu X, Wu S, Liu R, Debord J, Lou X, Xiao Y, A Label-Free Aptamer-Fluorophore Assembly for Rapid and Specific Detection of Cocaine in Biofluids, Anal. Chem 86 (2014) 11100–11106. doi: 10.1021/ac503360n. [DOI] [PubMed] [Google Scholar]
- [72].Pei R, Stojanovic MN, Study of thiazole orange in aptamer-based dye-displacement assays, Anal. Bioanal. Chem 390 (2008) 1093–1099. doi: 10.1007/s00216-007-1773-2. [DOI] [PubMed] [Google Scholar]
- [73].Ji D, Wang H, Ge J, Zhang L, Li J, Bai D, Chen J, Li Z, Label-free and rapid detection of ATP based on structure switching of aptamers, Anal. Biochem 526 (2017) 22–28. doi: 10.1016/j.ab.2017.03.011. [DOI] [PubMed] [Google Scholar]
- [74].Wang J, Jiang Y, Zhou C, Fang X, Aptamer-Based ATP assay using a luminescent light switching complex, Anal. Chem 77 (2005) 3542–3546. doi: 10.1021/ac050165w. [DOI] [PubMed] [Google Scholar]
- [75].Pei R, Rothman J, Xie Y, Stojanovic MN, Light-up properties of complexes between thiazole orange-small molecule conjugates and aptamers, Nucleic Acids Res. 37 (2009) e59. doi: 10.1093/nar/gkp154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yang K-A, Pei R, Stefanovic D, Stojanovic MN, Optimizing cross-reactivity with evolutionary search for sensors, J. Am. Chem. Soc 134 (2012) 1642–1647. doi: 10.1021/ja2084256. [DOI] [PubMed] [Google Scholar]
- [77].Yang K-A, Chun H, Zhang YM, Pecic S, Nakatsuka N, Andrews AM, Worgall TS, Stojanovic MN, High-affinity nucleic-acid-based receptors for steroids, ACS Chem. Biol 12 (2017) 3103–3112. doi: 10.1021/acschembio.7b00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Syu Y-C, Hsu W-E, Lin C-T, Review—Field-effect transistor biosensing: Devices and clinical applications, ECS J. Solid State Sci. Technol 7 (2018) Q3196–Q3207. [Google Scholar]
- [79].Shoorideh K, Chui CO, On the origin of enhanced sensitivity in nanoscale FET-based biosensors, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 5111–5116. doi: 10.1073/pnas.1315485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kim J, Rim YS, Chen H, Cao HH, Nakatsuka N, Hinton HL, Zhao C, Andrews AM, Yang Y, Weiss PS, Fabrication of high-performance ultrathin In2O3 film field-effect transistors and biosensors using chemical lift-off lithography, ACS Nano. 9 (2015) 4572–4582. doi: 10.1021/acsnano.5b01211. [DOI] [PubMed] [Google Scholar]
- [81].Weiss PS, Trevor PL, Cardillo MJ, Gas–surface interactions on InP monitored by changes in substrate electronic properties, J. Chem. Phys 90 (1989) 5146–5153. doi: 10.1063/1.456557. [DOI] [Google Scholar]
- [82].Vacic A, Criscione JM, Rajan NK, Stern E, Fahmy TM, Reed MA, Determination of molecular configuration by debye length Modulation, J. Am. Chem. Soc 133 (2011) 13886–13889. doi: 10.1021/ja205684a. [DOI] [PubMed] [Google Scholar]
- [83].Nakatsuka N, Yang K-A, Abendroth JM, Cheung KM, Xu X, Yang H, Zhao C, Zhu B, Rim YS, Yang Y, Weiss PS, Stojanovic MN, Andrews AM, Aptamer–field-effect transistors overcome Debye length limitations for small-molecule sensing, Science. 362 (2018) 319–324. doi: 10.1126/science.aao6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Park J-W, Tatavarty R, Kim DW, Jung H-T, Gu MB, Immobilization-free screening of aptamers assisted by graphene oxide, Chem. Commun 48 (2012) 2071–2073. doi: 10.1039/c2cc16473f. [DOI] [PubMed] [Google Scholar]
- [85].Gu H, Duan N, Wu S, Hao L, Xia Y, Ma X, Wang Z, Graphene oxide-assisted non-immobilized SELEX of okdaic acid aptamer and the analytical application of aptasensor, Sci. Rep 6 (2016) 21665. doi: 10.1038/srep21665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Quang NN, Perret G, Ducongé F, Applications of high-throughput sequencing for in vitro selection and characterization of aptamers, Pharmaceuticals. 9 (2016) 76. doi: 10.3390/ph9040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Röthlisberger P, Hollenstein M, Aptamer chemistry, Adv. Drug Delivery Rev 134 (2018) 3–21. doi: 10.1016/j.addr.2018.04.007. [DOI] [PubMed] [Google Scholar]