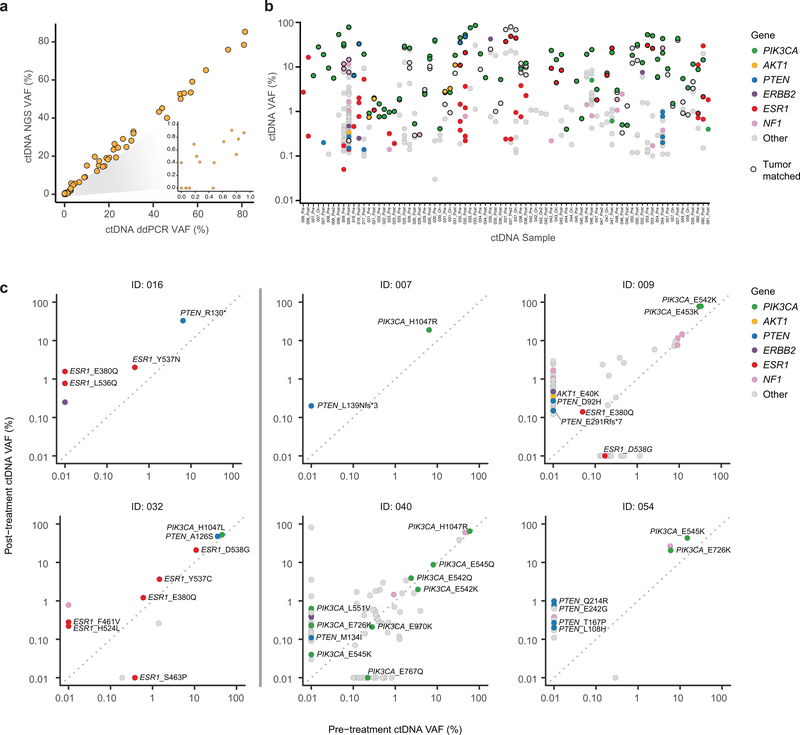
Extended Data Fig. 1. Characterization of ctDNA variants.
a) Comparison of variant allele fraction (VAF) of PIK3CA mutations measured using the targeted ctDNA assay (y-axis) and ddPCR (x-axis). cfDNA samples extracted from 65 samples (35 patients) with canonical hotspot PIK3CA mutations were subjected to ddPCR. An aliquot of the same cfDNA isolate was used for targeted DNA assay (G360 assay, Guardant Health, CA). b) ctDNA VAFs of all the somatic variants detected in ctDNA restricted to 38 patients with available tumor next generation sequencing results. Colors indicate the altered gene and black borders indicate whether the alteration was detected in the tumor tissue. c) Comparison of VAF of mutations detected in the pre-treatment and post-treatment ctDNA samples of six patients with evaluable paired ctDNA specimens and PTEN loss-of-function mutations in either sample. The colors of the circles indicate mutated gene.