Figure 3.
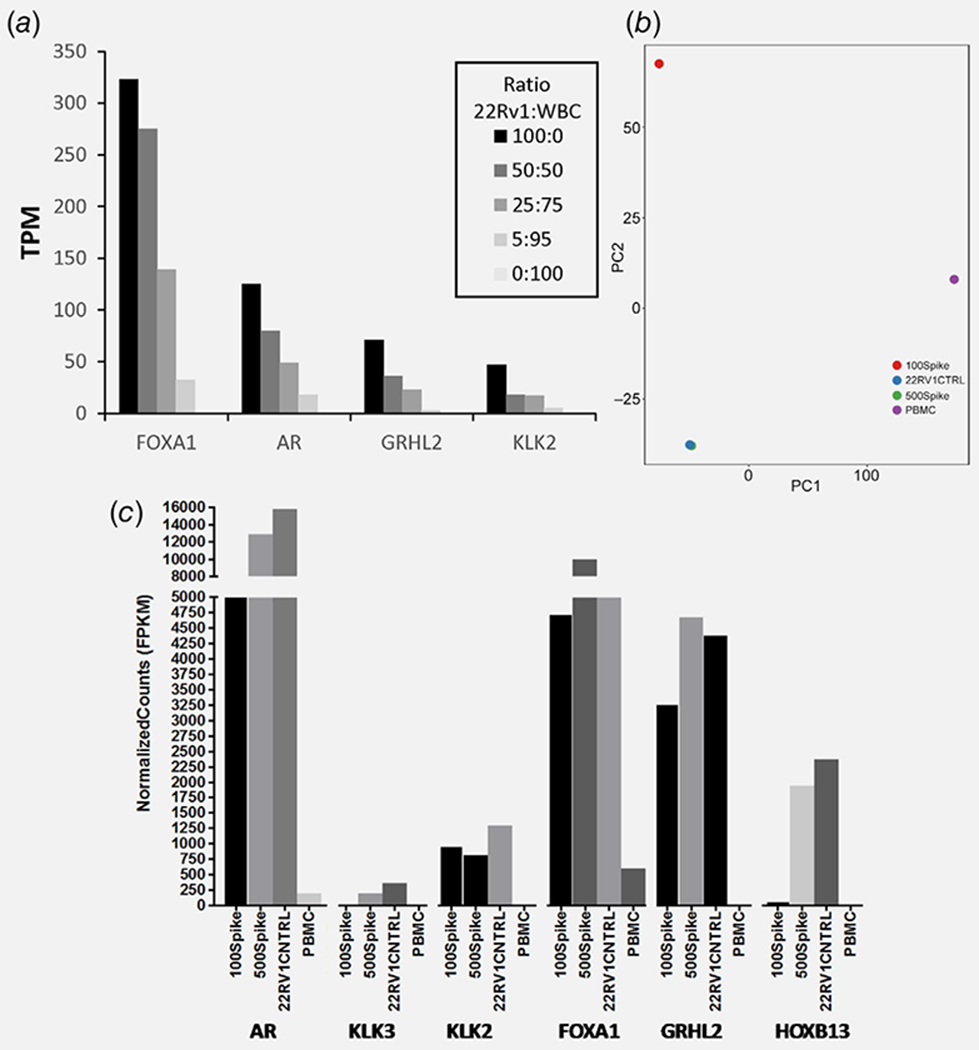
Feasibility of low-input RNA-seq analysis using cell line engineered controls as surrogates for PC patient samples. (a) 22Rv1 RNA and WBC RNA were mixed at the ratios specified, each representing different numbers of CTC equivalents. Each sample equaled 1 ng total RNA (~100 cell) and was used for low-input library prep and RNA-seq, and the PC-specific transcripts were quantified. (b) 22Rv1 cells (100 and 500 cells) were spiked into 7.5 ml healthy donor blood prior to enrichment, low-input library prep and RNA-seq. PCA discriminating 22Rv1 spiked-in samples from bulk PBMCs and cell line control is shown. (c) PC-specific transcript levels quantified from the RNA-seq data for these engineered blood samples. [Color figure can be viewed at wileyonlinelibrary.com]
