Fig. 3.
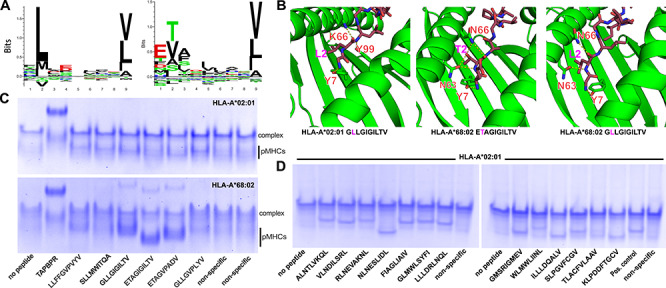
Specificity of peptide loading on CHO-derived HLA-A*02:01 and HLA-A*68:02. (A) Sequence logos of 9 mer epitopic peptides (IEDB database https://www.iedb.org), ranked by frequency (Seq2logo https://doi.org/10.1093/bioinformatics/btp033). (B) Structure models of peptides GLLGIGILTV, ETAGIGILTV and GLLGIGILTV bound to HLA-A*02:01 and HLA-A*68:02. We used a comparative modeling approach in the program Rosetta, with templates obtained from X-ray structures with PDB accession codes 4JFO, 3MRK and 4JFF, respectively (Nerli and Sgourakis, 2020). The dashed yellow lines indicate polar contacts. (C) Native gel shift assay showing pMHC-I formation in the presence of a panel of peptides with predicted affinities for HLA-A*02:01 and HLA-A*68:02 shown in Supplementary Table S1. Nonspecific peptides are YPNVNIHNF and RGPGRAFVTI, respectively (D) Same assay as in (C) showing peptide binding on HLA-A*02:01 for a panel of SARS-CoV-2 epitopic peptides predicted using a structure-based algorithm (Nerli and Sgourakis, 2020) (Supplementary Table S1). As a positive control (pos. control), we used the confirmed epitopic peptide NLVPMVATV from CMV, and as a negative control a nonspecific peptide with sequence YPNVNIHNF (NIH p29). Native gels (12% polyacrylamide) were electrophoresed at 90 V for 5 h at 4°C before visualization with InstantBlue (Expedeon).
