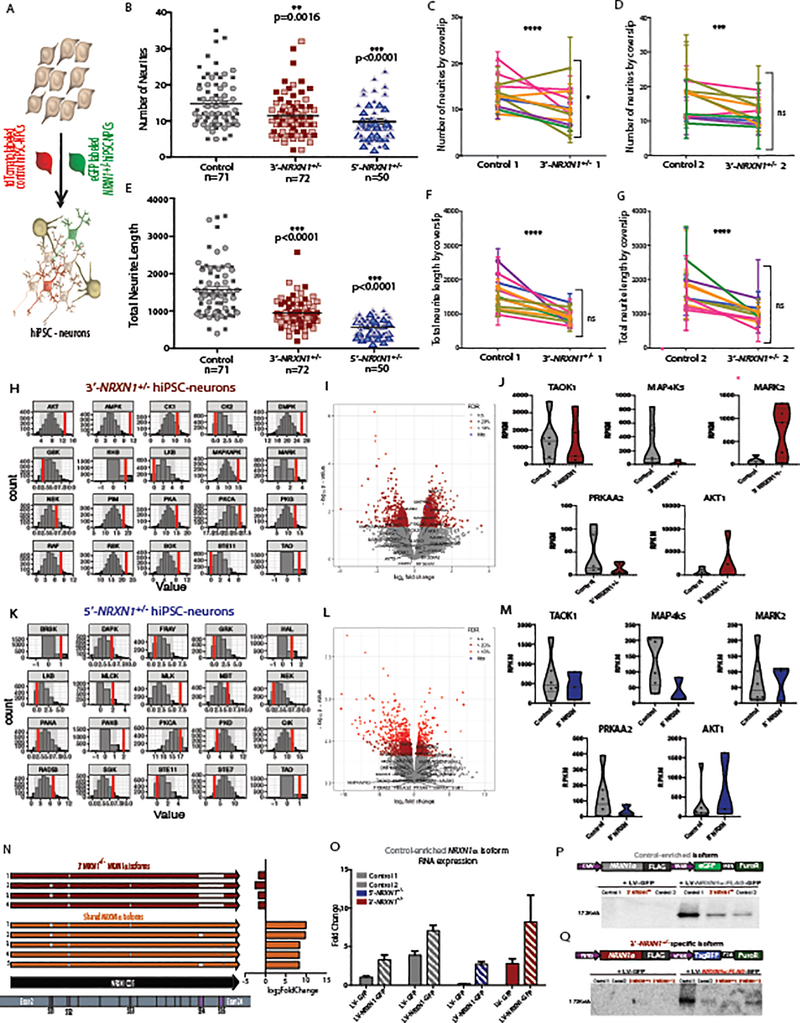
Extended Data Fig. 8. Investigation of hiPSC-neuron morphology, cellular signaling, and NRXN1 overexpression.
a, Strategy to label individual hiPSC-neurons. b-g, Mean neurite number across genotypes (control 71 neurons, 2 donors; 3’-NRXN1+/− 72 neurons, 2 donors; 5’-NRXN1+/− 50 neurons 2 donors) “****” indicates P < 0.00001 and “***”indicates P < 0.0001 by one-way ANOVA with Holm-Sidak’s test; (b) or by coverslip (2 donors, 12 coverslips, 3 regions each) (c,d) mean neurite length across genotypes (control 71 neurons, 2 donors; 3’-NRXN1+/− 72 neurons, 2 donors; 5’-NRXN1+/- 50 neurons, 2 donors); (e) or by coverslip (2 donors, 12 coverslips, 3 regions each) (f,g). Two donors per genotype indicated by different shading within each plot. h, Differentially active kinases (3’- NRXN1+/− hiPSC-neurons: 6 samples, 2 donors; controls: 5 samples, 2 donors). i, Volcano plot of –log10(P-value) and log2(FoldChange) from linear model of RNA-seq (3’-NRXN1+/− hiPSC-neurons: 5 samples, 2 donors; controls 6 samples, 2 donors); DE kinase associated genes labeled. j, Violin plot of median and quartiles of RPKM for kinase hits with largest fold-change in RNA-seq (3’-NRXN1+/− hiPSC-neurons: 5 samples, 2 donors; controls: 6 samples, 2 donors). k, Differentially active kinases in (5’- NRXN1+/− hiPSC-neurons: 3 samples, 1 donors; controls 5 samples, 2 donors). l, Volcano plot of –log10(P-value) and log2(FoldChange) from linear model of RNA-seq (5’-NRXN1+/− hiPSC-neurons: 3 samples, 1 donors; controls 6 samples, 2 donors); DE kinase associated genes labeled. m, Violin plot of median and quartiles of RPKM for kinase hits with largest fold change values in RNA-seq (5’-NRXN1+/− hiPSC-neurons: 3 samples, 1 donors; controls 6 samples, 2 donors). n, Isoform constructs for overexpression with log2(FoldChange) from hybrid sequencing dataset. o, Mean fold-change from qPCR of NRXN1 expression (3 replicates per condition. p, Representative western blot (2 replicates) for anti-FLAG (48hr expression of control-enriched NRXN1α-FLAG). q, Representative western blot (2 replicates) for anti-FLAG (48hr expression of 3’-NRXN1+/− specific NRXN1α-FLAG). All error bars are s.e.