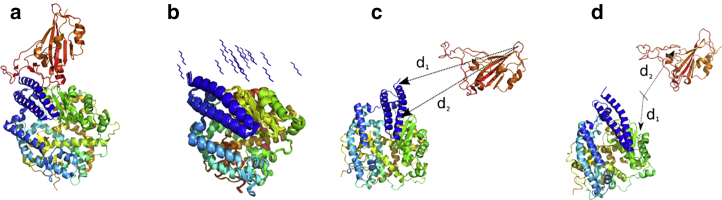
Figure 1.
(a) Crystal structure of the SARS CoV-2 spike protein CoV-2 RBD bound with ACE2 (PDB: 6M0J (19)). ACE2 is shown in blue, cyan, yellow, and green, and CoV-2 RBD is shown in orange. (b) 20 different hexapeptide (YKYRYL) conformations are used as initial starting models of independent 50-ns equilibrium MD simulations in explicit solvent. (c) Shown is the starting structure of the enhanced CORE-MD simulation of CoV-2 RBD to ACE2. (d) Shown is the starting structure of the enhanced CORE-MD simulation of the same system in the presence of the hexapeptide. The distances d1 and d2 used in the free energy projections (see Fig. 4) are indicated in (c) and (d). To see this figure in color, go online.