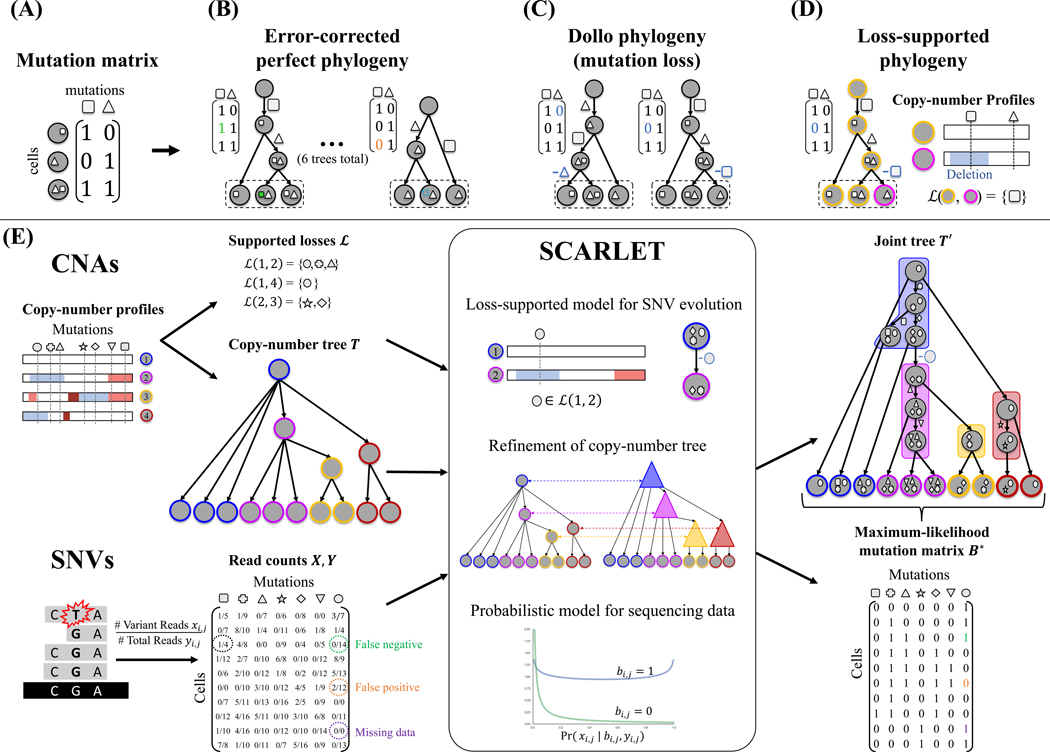
Figure 1: Loss-supported phylogeny model, and SCARLET algorithm for the Maximum Likelihood Loss-Supported Refinement Problem.
(A) A mutation matrix with two mutations in three cells does not admit a perfect phylogeny. This may be due to either errors or mutation losses. (B) Under the infinite sites model, existing methods correct errors in the observed matrix to yield a perfect phylogeny. (C) Under the Dollo model, existing methods identify mutation losses to explain violations of the infinite sites model. Both the infinite sites and Dollo models yield multiple equally-plausible phylogenies. (D) The loss-supported model overcomes this ambiguity by using copy-number data to constrain mutation losses. (E) SCARLET algorithm for the Maximum Likelihood Loss-Supported Refinement problem. SCARLET integrates single-nucleotide variants (SNVs) and copy-number aberrations (CNAs) for tumor phylogeny inference. For CNAs, observed copy-number profiles indicate amplified (red) or deleted (blue) genomic regions along the entire genome and are used to obtain two inputs for SCARLET. First, supported loss sets ℒ(c, c′) for pairs of copy-number profiles (empty sets are not shown) indicate mutations that are affected by deletions. Second, a copy-number tree T which describes the ancestral relationships between observed cells (leaves) as determined by copy-number profiles. For SNVs, variant X and total Y read counts are provided to SCARLET for every cell and every mutation. SCARLET computes a joint tree T′ on the observed cells and a maximum-likelihood mutation matrix B* by constraining mutation losses to the supported loss sets ℒ, computing a refinement T′ of T, and selecting the maximum-likelihood B* using a probabilistic model for the presence (bi,j = 1) or absence (bi,j = 0) of each SNV in each cell.