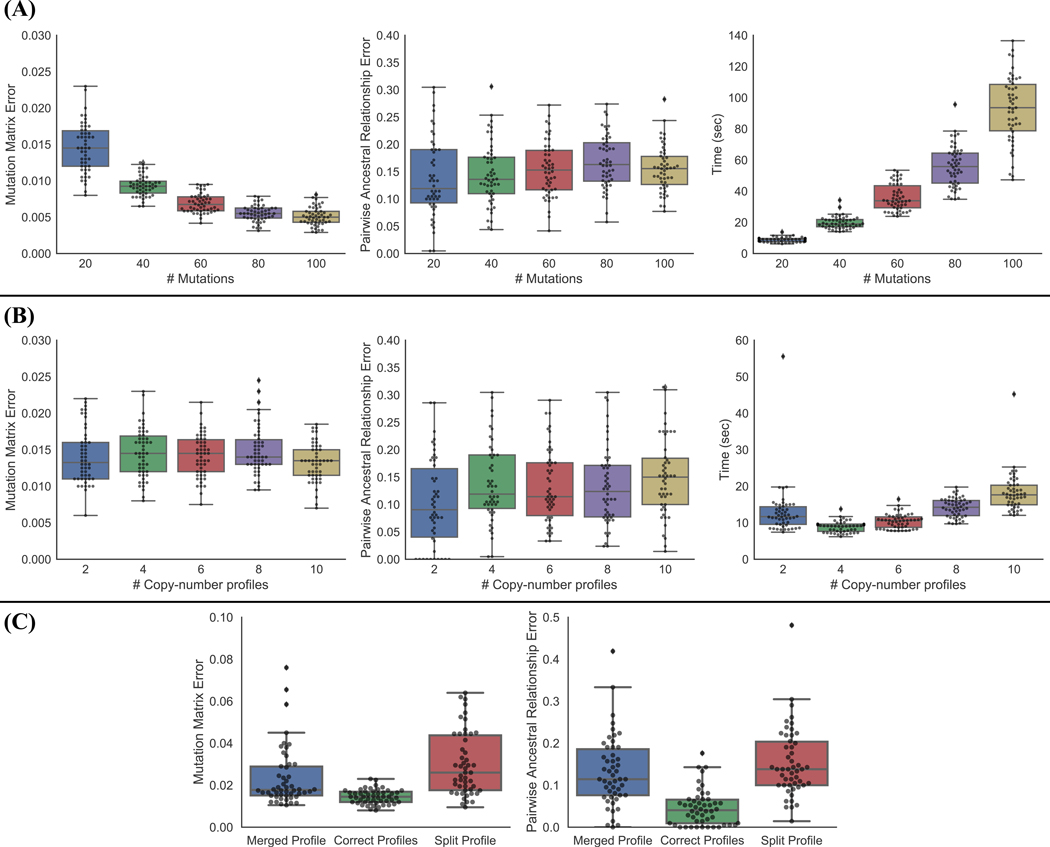
Figure 3: SCARLET scales to larger datasets and tolerates errors in copy-number profiles.
(A) SCARLET results on simulated data with varying number of mutations, n = 100 cells and k = 4 copy-number profiles. (Left) Mutation matrix error; (Center) pairwise ancestral relationship error; (Right) runtime. (B) SCARLET results on simulated data with varying number of copy-number profiles, n = 100 cells and m = 20 mutations. (Left) Mutation matrix error; (Center) pairwise ancestral relationship error; (Right) runtime. (C) SCARLET results on simulated data with incorrect number of copy-number profiles. We introduce errors in the number of copy-number profiles by either merging two distinct copy-number profiles together (“Merged Profile”), or splitting one copy-number profile into two (“Split Profile”) and compare performance against the correct number of copy-number profiles.