FIGURE 6.
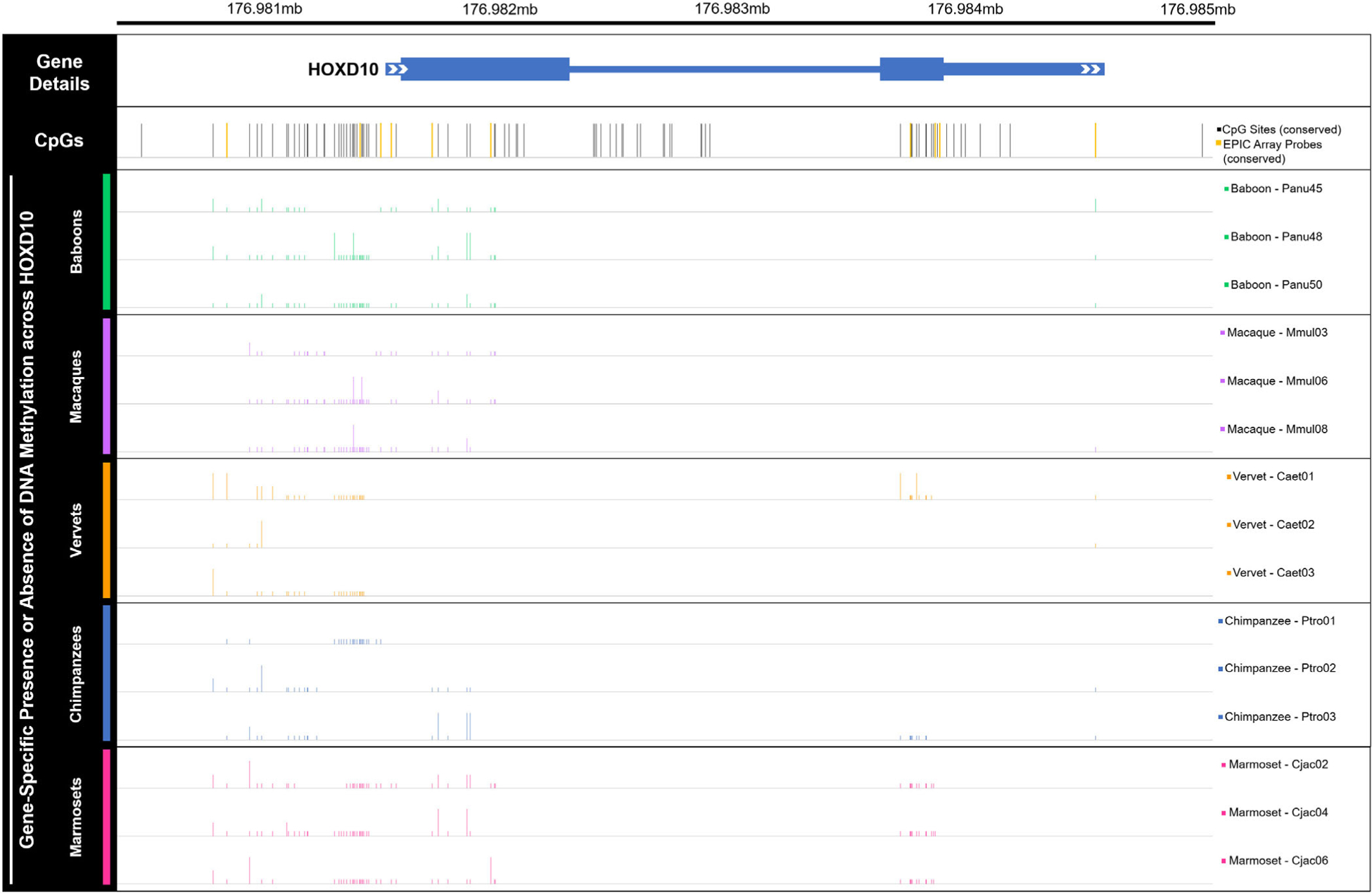
Gene-specific methylation levels across HOXD10 in nonhuman primates. Bar plot of DNA methylation across the HOXD10 gene (hg19 chr2:176,981,492–176,984,670), as well as upstream and downstream several hundred bases (hg19 chr2:176,980,532–176,985,117). Bars depict the presence (tall bar), partial presence (medium bar), or absence (low bar) of methylation at human derived CpG sites in 15 nonhuman primate samples—3 baboons, 3 macaques, 3 vervets, 3 chimpanzees, and 3 marmosets. While regular sequencing was very successful, bisulfite sequencing was less successful, with several sequence reads uninterpretable. As such, nonhuman primate methylation data are only available for a subset of the CpGs known in humans. Partial presence of methylation was called when sequencing fluorescence peaks for cytosine and thymine were both present at a particular site and one was at least half the size of the other. Overall, these data provide additional information regarding gene-specific methylation levels across HOXD10. CpG sites that were also targeted by the EPIC array are highlighted in yellow and include cg18115040 (chr2, position 176,981,328), cg25371634 (chr2, position 176,981,422), cg13217260 (chr2, position 176,981,469), cg03918304 (chr2, position 176,981,654), cg17489939 (chr2, position 176,981,919), cg26708100 (chr2, position 176,983,815), cg10393811 (chr2, position 176,983,927), cg08992581 (chr2, position 176,983,949), and cg06005169 (chr2, position 176,984,634). See Table S18 and Files S8 and S9 for additional information
