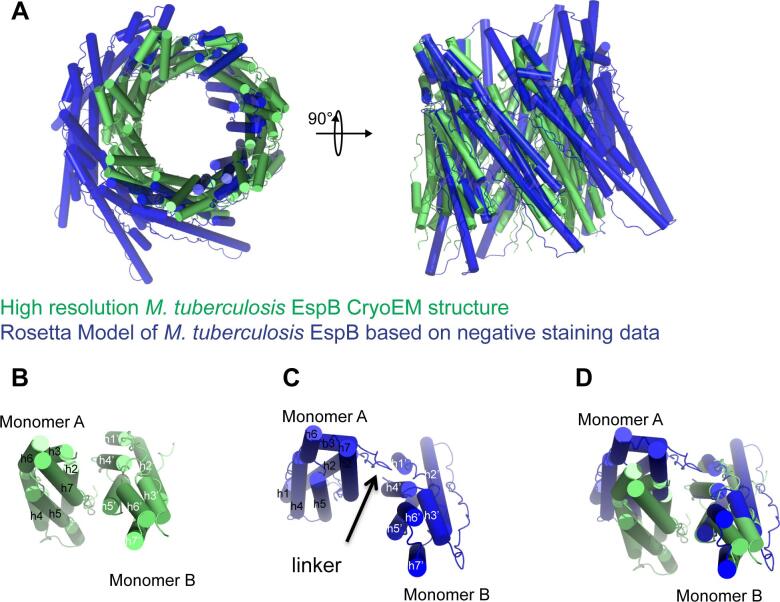
Fig. 3.
Structural comparison between the high-resolution M. tuberculosis EspB cryoEM structure and the previously published Rosetta model of M. tuberculosis EspB based on negative staining data (3 J83). A) Superimposition of the two heptamers on chain A in top and side views. B) Interfaces between two subunits in the cryoEM structure. C) Interfaces between two subunits in the Rosetta model based on negative staining data. D) Comparison of the interfaces between monomers in the two structures. High resolution structure allows to identify residues present at the interface.