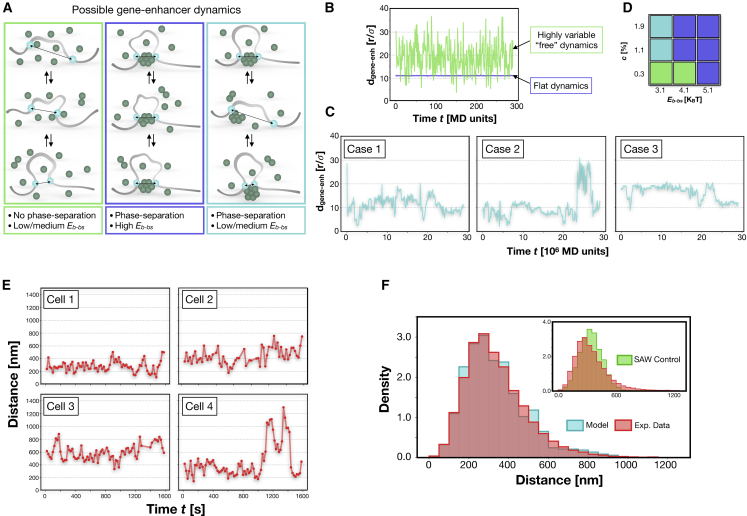
Figure 3.
The PS process produces different regimes of gene-enhancer contact dynamics. (A) The affinity between bs and binders influences the gene-enhancer contacts in time, and different dynamics emerge. (B) An example of gene-enhancer distance dynamics in which a stable contact is not formed (green curve, Eb-bs = 3.1 KBT, Eb-b = 3.0 KBT, and c is ∼0.3%, and there is no phase-separated cluster) and in which a highly stable contact (blue curve, Eb-bs = 5.1 KBT, Eb-b = 3.0 KBT, and c is ∼1.1%) mediated by a phase-separated cluster is formed. (C) Three examples of contact dynamics for intermediate affinity Eb-bs = 3.1 KBT (Eb-b = 3.0 KBT). Cases 1 and 2 are with c ∼1.1%, and case 3 is with c ∼1.9%. Here, the contact is stable, yet results are much more variable because the bs can move on the surface of the cluster and can also detach. (D) Diagram summarizing the different dynamic regimes and their corresponding parameters. (E) Examples of experimental single-cell gene-enhancer dynamics from Sox2 and its superenhancer SCR in mouse embryonic stem cells. Data are taken from (33). The profile results are very similar to the simulated dynamics in (C). (F) Comparison between the model (cyan) and experimental (red) distributions of distances. The distributions are consistent (Kolmogorov-Smirnov KS test, p-value > 0.01). The inset shows the comparison with the control distribution of the free SAW case (KS test, p-value = 10−24). To see this figure in color, go online.