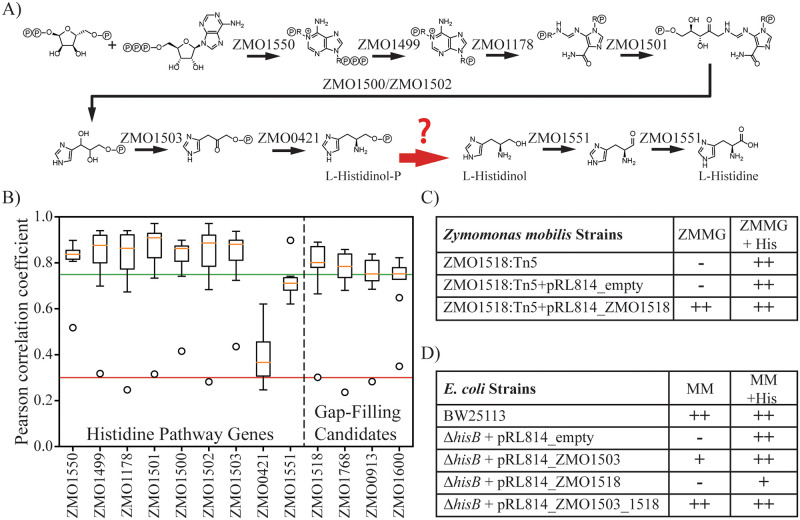
Fig 3. Identification of the histidinol-phosphatase gene.
(A) An overview of the histidine biosynthesis pathway, converting 5-phosphoribosyl diphosphate (PRPP) to L-histidine. Note that ZMO1500 and ZMO1502 are subunits associated with the same reaction. Highlighted in red is the gap-filled histidinol-phosphatase reaction lacking an annotated gene. (B) Boxplot of the cofitness values for genes with known histidine biosynthesis genes. Cofitness of the genes in the histidine pathway with the eight genes of the histidine pathway are shown on the left. Candidate genes for the histidinol-phosphatase reaction (i.e., those with the highest average cofitness to the known genes) are shown on the right. The low cofitness outlier in all cases (except ZMO1551) corresponds to the cofitness with the ZMO0421 gene. (C) Growth experiments showed the ZMO1518::Tn5 mutant was a histidine auxotroph that could be rescued by complementation with ZMO1518 on a plasmid. Phenotypes are categorized as growth (++), weak growth (+), and no growth (-). (D) Growth experiments in E. coli ΔhisB demonstrate that ZMO1518 encodes a histidinol-phosphatase. Note that in E. coli, hisB has two functions catalyzing both the sixth and eighth steps in histidine biosynthesis, and therefore the E. coli ΔhisB knockout requires complementation with both ZMO1503 and ZMO1518.