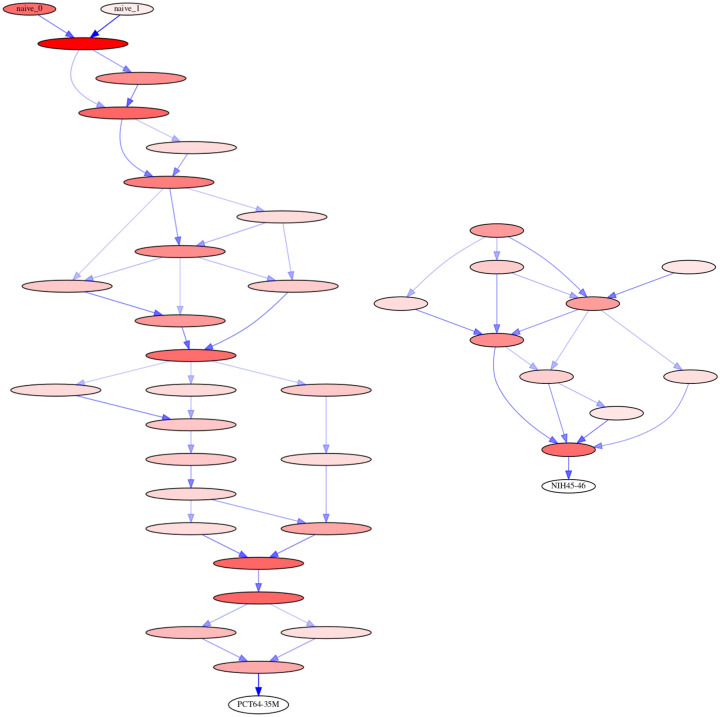
Fig 4. Naive-to-tip sequence trajectory graphics.
The linearham-inferred naive-to-tip amino acid sequence trajectories for the pruned PC64 dataset of 100 sequences and the trimmed VRC01 alignment of 268 sequences, displaying only the edges that satisfy the given posterior probability threshold, and only the nodes that contact edges above the threshold. The tip sequences of interest for the PC64 and VRC01 datasets are chosen to be PCT64-35M and NIH45-46, respectively, and we use 0.04 probability cutoffs for these lineage graphics (such that any edge with probability less than this threshold is discarded). The nodes correspond to unique ancestral sequences filled with red color, where the opacity is proportional to the posterior probability of the associated sequence. The directed edges connecting nodes represent ancestral sequence transitions and are shaded blue with an opacity proportional to the posterior probability of the associated sequence transition. Nodes without any probable edges connecting them are not displayed in these graphics. The absence of many nodes for VRC01 indicates that these naive-to-tip sequence trajectories are highly uncertain. A more detailed version of this graphic, including predicted lineage mutations, is included as S1 Fig.