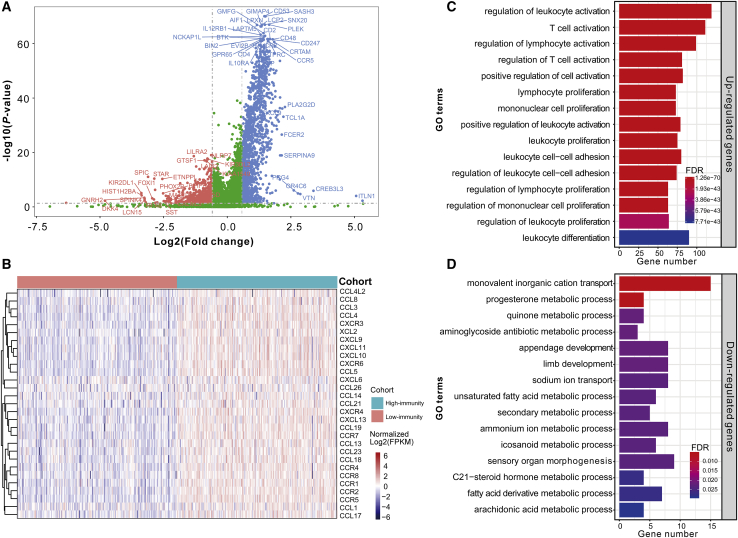
Figure 2.
Investigation of the Immune Infiltration-Dependent Expression Change
(A) Volcano plot showing the differentially upregulated (blue nodes) and downregulated genes (red nodes). (B) Heatmap displaying the expression changes of 29 chemotactic factors. The colors of the top bar and central plot represent the grouping information and gradient of normalized log2FPKM (fragments per kilobase of transcript per million mapped reads), respectively. (C) Bar plot showing the biological processes enriched by the upregulated genes. (D) Bar plot showing the biological processes enriched by the downregulated genes. The y axis in (C) and (D) reflects the name of GO terms. The x axis reflects the overlapped gene numbers between each GO term and query gene set. The color of the bars represents the gradient of adjusted p values (FDR correction).