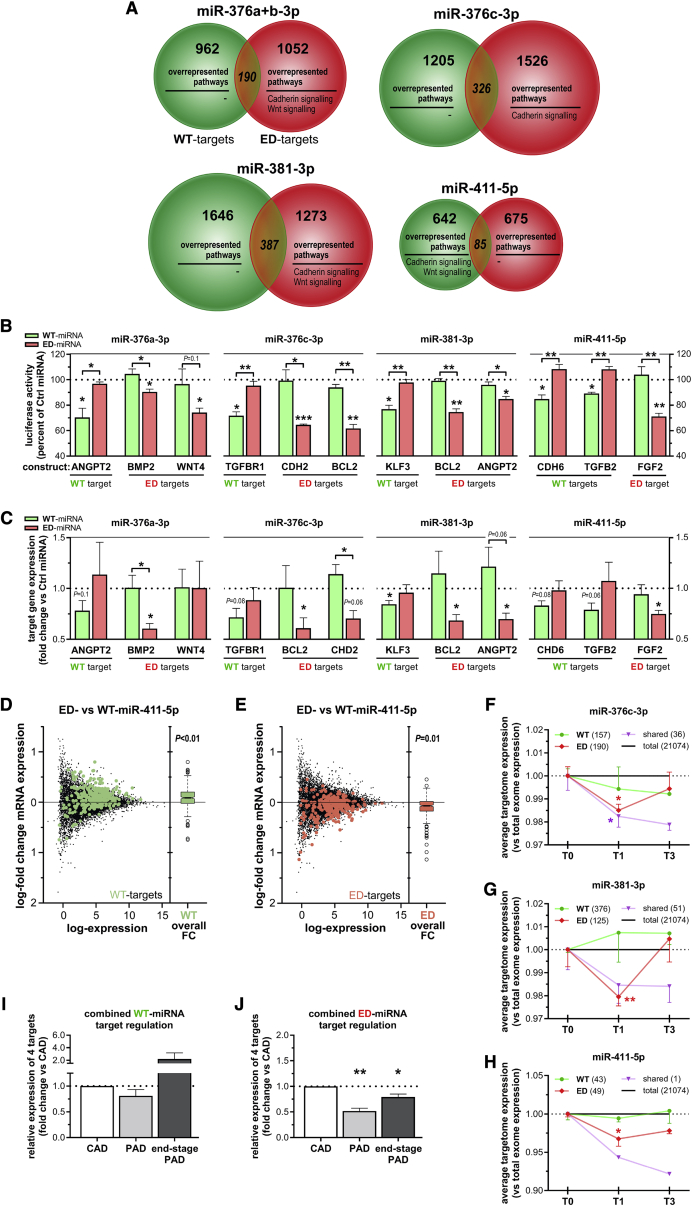
Figure 7.
Targetome Predictions and Validation of Target Sequence Binding and Endogenous Target Regulation
(A) Venn diagram of putative targetomes for WT-microRNA (green) and ED-microRNA (red) representing the putative targets that all three prediction algorithms indicated to be targeted. Within each putative targetome, significantly enriched pathways were displayed (see Table S4). (B) To examine the change in target binding induced by editing, luciferase assays were performed with endogenous microRNA binding sequences from vasoactive targets for either a WT-microRNA or a ED-microRNA as indicated. MicroRNA-specific target regulation was assessed by examining luciferase reporter activity after co-transfecting HeLa cells with a WT-microRNA mimic (green) or a ED-microRNA mimic (red), normalized to transfection of a non-targeting microRNA mimic (Ctrl microRNA). (C) Endogenous target regulation within HUAFs after transfection-mediated overexpression of microRNA mimics as indicated. For (B) and (C), data shown represent the averages of three independent experiments and are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by a one-sample t test versus Ctrl microRNA or a two-sided Student’s t test to compare WT-microRNA versus ED-microRNA treatments. (D and E) The log fold changes (FCs) of mRNA expression calculated from RNA-seq data obtained after overexpression of ED-miR-411-5p compared to overexpression of WT-miR-411-5p in HUAFs. Data represent averages of three independent experiments and are visualized using mean-difference plots, highlighting the predicted target genes (with a 0.5 binding score threshold to minimize false positives; see Table S6) that were uniquely targeted by the WT-miR-411-5p (green, D) or ED-miR-411-5p (red, E). The overall distribution of the logFC per targetome is shown in the boxplots. Differential expression of each targetome was tested using ROAST.62Figure S6 shows target gene regulation by WT-miR-411-5p and ED-miR-411-5p compared to a non-targeting control microRNA. (F–H) Analysis of conserved targetome expression with a predicted binding score of >0.5 (see Table S7) in whole-muscle tissue of C57BL/6 mice subjected to HLI using whole-genome expression data obtained via microarray.21 Mean fold change in targetome expression of WT, ED, or shared targets (green, red, and purple, respectively) of miR-376c-3p (F), miR-381-3p (G), and miR-411-5p (H) targets after HLI. Per targetome, the number of conserved targets that were detected above array background levels is indicated between brackets. Targetome expression was normalized to expression of total number of genes detected (black line) and presented as mean ± SEM of at least three different mice per time point. ∗p < 0.05, ∗∗p < 0.01, versus whole-genome expression by a two-sided Student’s t test. (I and J) The combined average mRNA expression of the WT-microRNA targets (I) or ED-microRNA targets (J) validated in (B) and (C) within the LLV samples from three different patient groups (see Figure 6): CAD patients (LLVs without PAD, n = 8), PAD patients (LLVs suffering severe PAD, n = 6), and end-stage PAD patients (LLVs suffering end-stage PAD, n = 8). Combined average expression levels were calculated using the average expression of individual target genes shown in Figure S7 and are expressed as fold change of the CAD group. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, by a one-sample t test.