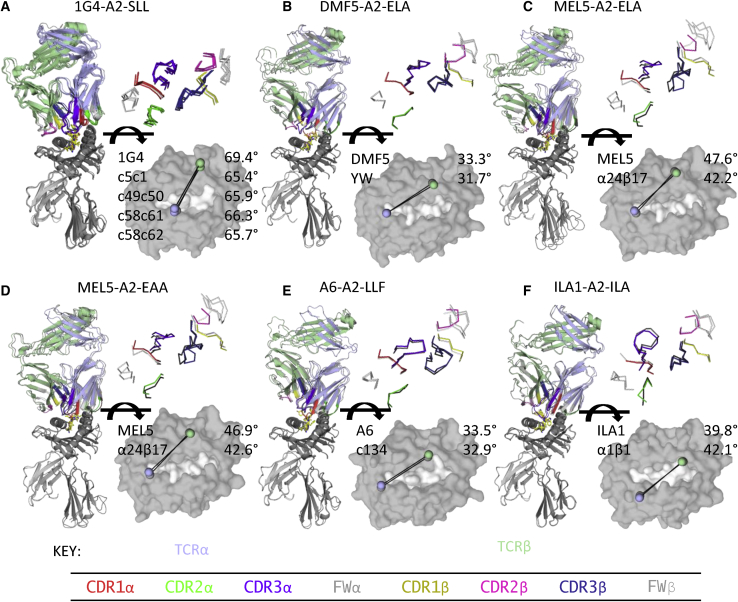
Figure 1.
Structural Comparison of Overall Binding Modes of Affinity Enhanced and Wild-Type TCR-pHLA Complexes Shows Virtually Identical Conformations
(A–F) Left: structural overlay of wild-type (WT) TCR-pHLA complexes versus affinity-enhanced (ae)TCR-pHLA complexes. TCR and HLA are shown as cartoon and peptide is shown as stick representation. Right top: overlay of CDR loop positions of WT and aeTCRs. Backbone locations of each CDR loop shown as line representation. In each panel, the WT TCR complex structure is in grayscale and the aeTCR complex structure is colored by chain: TCRα, blue; TCRβ, green; HLA, dark gray; β2m, light gray. Right bottom: TCR crossing angle of WT TCR-pHLA complexes versus aeTCR-pHLA complexes. pHLA is shown as surface representation (gray). TCRα (blue) and TCRβ (green) centroid locations of aeTCR structure are shown as spheres. (A) 1G4 TCR and multiple aeTCR variants in complex with HLA-A∗0201-SLLMWITQC. (B) DMF5 TCR and aeTCR variant DMF5_YW in complex with HLA-A∗0201-ELAGIGILTV. (C) MEL5 TCR and aeTCR variant MEL5_α24β17 in complex with HLA-A∗0201-ELAGIGILTV. (D) MEL5 TCR and aeTCR variant MEL5_α24β17 in complex with HLA-A∗0201-EAAGIGILTV. (E) A6 TCR and aeTCR variant A6_c134 in complex with HLA-A∗0201-LLFGYPVYV. (F) ILA1 TCR and aeTCR variant ILA1_α1β1 in complex with HLA-A∗0201-ILAKFLHWL.