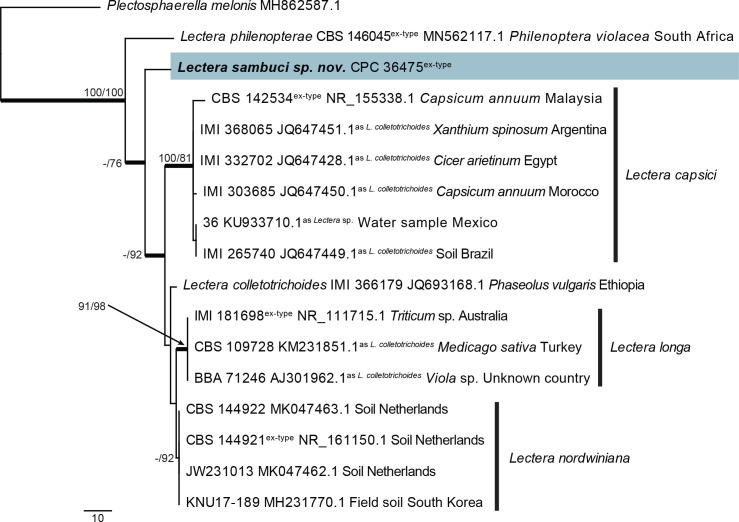
Fig. 30.
The first of 16 equally most parsimonious trees obtained from a phylogenetic analysis of the Lectera ITS alignment (17 strains including the outgroup; 476 characters including alignment gaps analysed: 371 constant, 80 variable and parsimony-uninformative and 25 parsimony-informative). The tree was rooted to Plectosphaerella melonis (strain CBS 489.96, GenBank MH862587.1) and the scale bar indicates the number of changes. Parsimony bootstrap (PBS) and neighbour joining bootstrap support (NJBS) values higher than 74 % are shown at the nodes (PBS/NJBS) and the treated species is highlighted with a coloured box and bold text. Species names are indicated to the right of the tree, or before the culture collection, GenBank accession numbers, substrate and country of origin. Type status and species name under which the sequence is deposited on GenBank are in superscript. Branches present in the strict consensus tree are thickened. Tree statistics: TL = 146, CI = 0.911, RI = 0.865, RC = 0.788.