Figure 6.
Transcriptional Identity of Hepatized Lymph Nodes (HLNs)
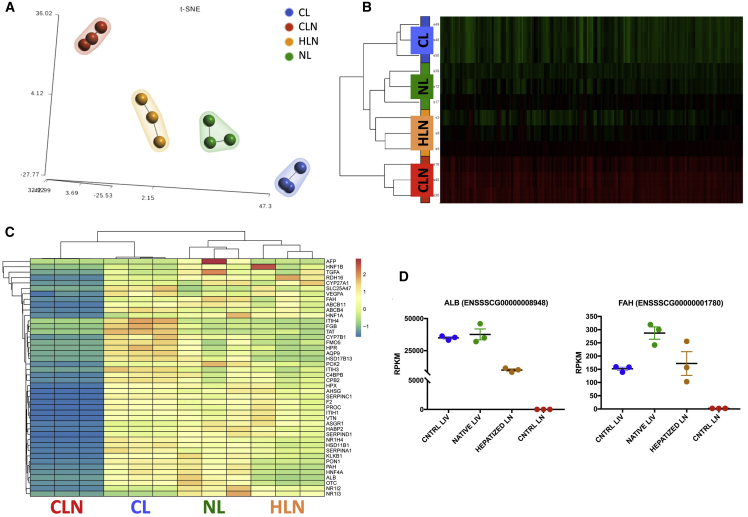
(A) t-Distributed Stochastic Neighbor Embedding (t-SNE) 2D plot of RNA-seq data, color-coded by tissue sample, showing evidence of HLN tendency to cluster with liver samples. CL, control liver (wild type); CLN, control lymph node (FAH knockout [KO]); NL, native liver (treated FAH KO). (B) Heatmap showing the normalized RNA-seq expression levels (reads per kilobase million [RPKM]) of differentially expressed genes across the four conditions. Red and green color intensity indicates gene upregulation and downregulation, respectively. (C) Heatmap showing the normalized RPKM of liver-specific differentially expressed genes across the four conditions. Spectral colors were used, with blue indicating low expression values, yellow indicating intermediately expressed genes, and red representing highly expressed genes. (D) RPKM values for albumin (ALB) and Fah in CL, NL, HLN, and CLN. Data are mean ± SEM.