Figure 6.
SHIP1 Undergoes a Conformational Change upon Allosteric Regulator Binding
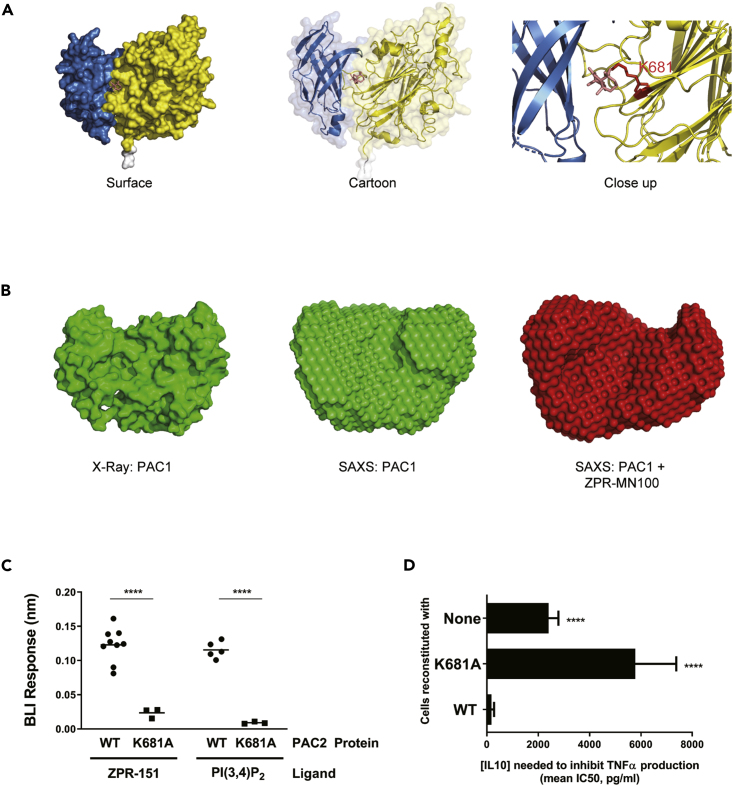
(A) Model of PAC2 based on crystallography data. The predicted binding pocket for the ZPR-MN100 (pink stick diagram) is located in the interface between C2 (blue) and phosphatase (yellow) domains. The binding pocket has amino acid residues K681A in close proximity with ZPR-MN100.
(B) SAXS model of PAC1 in the absence (apo-PAC1) and presence of (liganded) ZPR-MN100.
(C) Bio-layer interferometry (BLI) data of PAC2 WT and K681A loaded sensors exposed to either 20 μM of ZPR-151 or PI(3,4)P2. ∗∗∗∗p < 0.0001 comparing WT PAC2 and K681A (Unpaired Student's t test).
(D) TNFα production of 10 ng/mL LPS + IL10 stimulated cells reconstituted with WT or K681A SHIP1 or none (SHIP1 KO) determined by ELISA from which IC50 values for IL10 were calculated. ∗∗∗∗p < 0.0001 when compared with cells reconstituted with WT SHIP1 (Data represent means ± SD. Unpaired Student's t test). See also Figure S3 and Table S1.