Figure 2.
Most Selected Viral Populations Represent Rare and Uncharacterized Viral Sequences
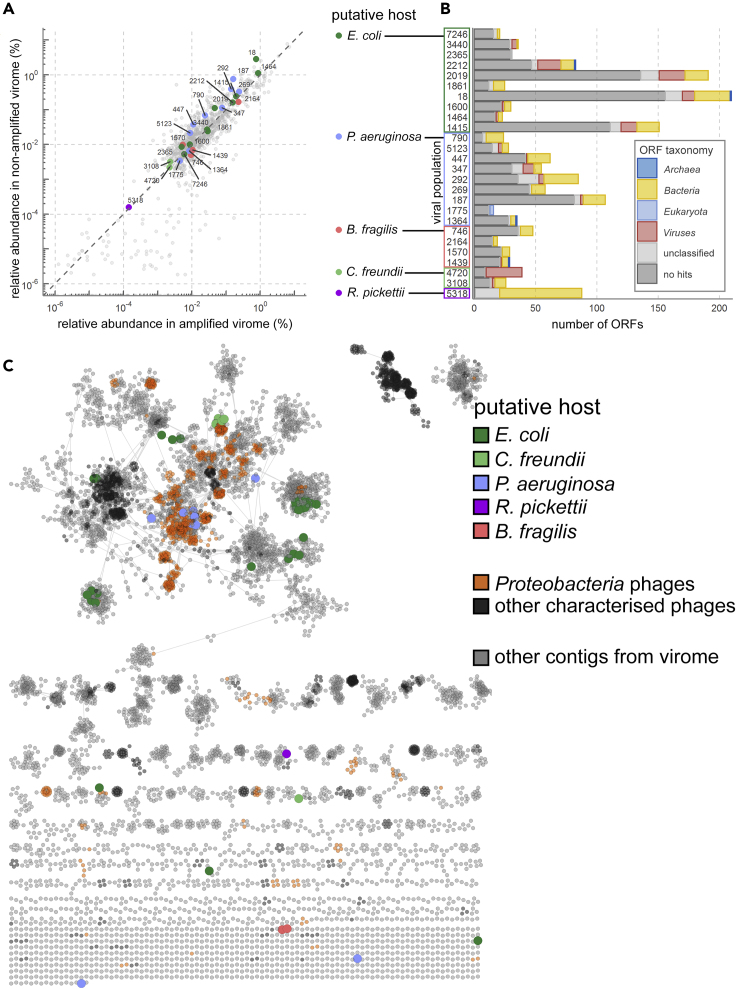
(A) Relative abundance of selected viral populations with adsorption predictions in the virome before and after MDA shows that AdsorpSeq is not biased for abundant or rare phage sequences. Numbers next to data points show the viral population number.
(B) ORF-level taxonomical predictions using CAT show most ORFs from selected viral populations have no similarities in the NCBI nr protein sequence database (dark gray). Some contigs had database hits but could not be classified because the hits involved proteins from different superkingdoms. These are labeled as unclassified (light gray).
(C) The hospital wastewater virome contained a large diversity of uncharacterized phage sequences, as shown by a gene-sharing network of 10,032 viral contigs and all phage genomes in the NCBI viral RefSeq database (Pruitt et al., 2007), made using vContact2 (Bin Jang et al., 2019). Large colored contigs represent those in the final selection of 26 putative adsorbing viral populations. Proteobacteria-infecting characterized phages are orange.