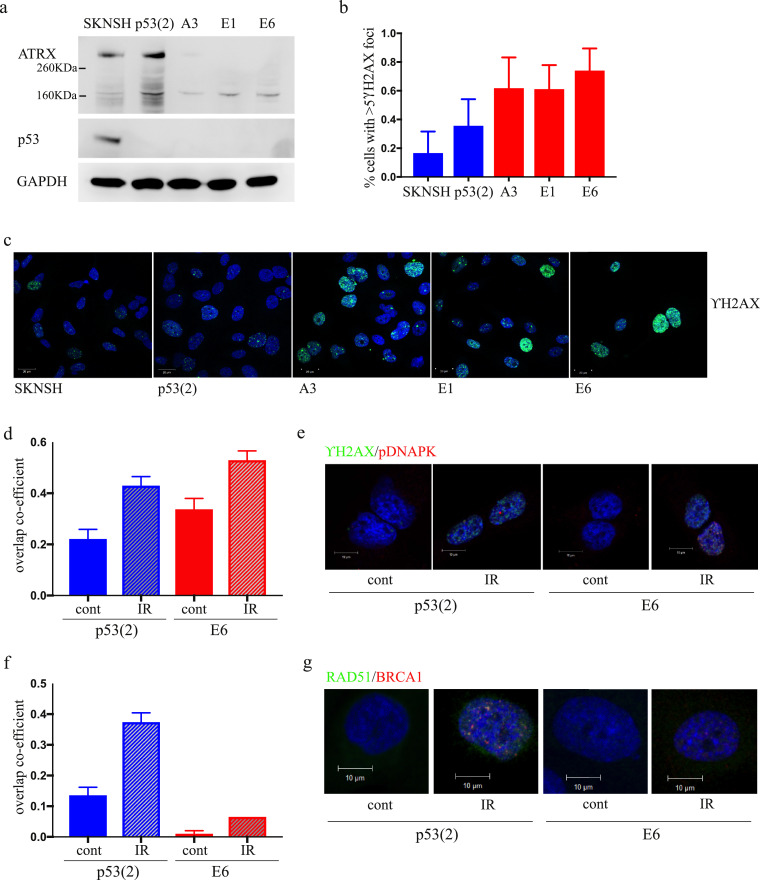
Fig. 1.
Generation of stable ATRX CRISPR Cas9 knock-out neuroblastoma cell lines results in increased DNA damage and HRR deficiency. (a) Western blot showing ATRX and p53 expression in the isogenic panel of cell lines generated by CRISPR Cas9 gene editing of SKNSH (b) Proportion of untreated cells with >5 γH2AX foci (error bars represent SD from 2 independent experiments, minimum 140 cells, p<0.0001 by one-way Anova). (c) Representative images showing γH2AX foci in untreated cells. (d) Quantification of the overlap coefficient of γH2AX and pDNAPK foci by immunofluorescence, 30 min post 10 Gy irradiation (IR) versus control (cont), in p53(2) and E6 cell lines (e) Representative images from pDNAPK (red) and γH2AX (green) co-localisation experiment. (f) Quantification of the overlap coefficient of BRCA1 and RAD51 foci by immunofluorescence, 8 hours post 10 Gy IR versus cont, in p53(2) and E6 cell lines. (g) Representative images from BRCA1 (red) and RAD51 (green) co-localisation experiment. For co-localisation experiments a minimum of 100 cells from 2 experiments were analysed by the ZEN software co-localisation analysis tool. Error bars represent SEM.