Abstract
A large set of multi-kinase inhibitors with high-confidence activity data was assembled and used to generate network representations revealing kinase relationships based upon shared inhibitors [1]. Compounds and activity annotations were originally selected from public repositories and organized in an in-house database from which the data set was extracted and curated. The new data set comprises more than 36,000 inhibitors with multiple activity annotations for a total of 420 human kinases (providing 81% coverage of the human kinome), representing a total of ∼127,000 kinase-inhibitor interactions. Use of the data is not limited to the network application reported in [1]. It can also be used, for example, for different types of compound promiscuity analysis or machine learning (such a multi-task modeling). In addition, the data set provides a large resource for complementing kinase drug discovery projects with external compound information.
Keywords: Human kinome, Multi-kinase inhibitors, Activity annotations, Compound-kinase interactions, Network representations
Specifications Table
| Subject | Drug discovery |
| Specific subject area | Computational analysis of compounds and activity data to explore inhibitor-based kinase relationships and identify representative kinases. |
| Type of data | Table Figure |
| How data were acquired | Data were acquired from a pre-established in-house database [2] and curated for applications using inhibitors with multi-kinase activity. |
| Data format | Secondary data Table (consistently formated) |
| Parameters for data collection | The following compound selection criteria were applied: (1) Inhibitors of human kinases, (2) multi-kinase activity, (3) valid SMILES representation [3], (4) standard potency measurements, (5) minimum potency of 1 µM. |
| Description of data collection | The source database of kinase inhibitors [2], from which the multi-kinase inhibitor data set reported herein was curated, was originally assembled from seven major compound repositories including ChEMBL [4], PubChem [5], Probes and Drugs Portal [6], BindingDB [7], PDBbind [8], ProteomicsDB [9], and Drug Target Commons [10]. |
| Data source location | Department of Life Science Informatics, B-IT, University of Bonn, Endenicher Allee 19c, d-53,115 Bonn, Germany. |
| Data accessibility | The data set is freely available for download from the public university cloud as a formated data file (csv format) via the following link: https://uni-bonn.sciebo.de/s/reyHRZXYW1D26sq |
| Related research article | O. Laufkoetter, S. Laufer, J. Bajorath, Identifying representative kinases for inhibitor evaluation via systematic analysis of compound-based target relationships, Eur. J. Med. Chem. 204 (2020) 112641. https://doi.org/10.1016/j.ejmech.2020.112641[1] |
Value of the Data
-
•
The large curated set of multi-kinase inhibitors with 81% coverage of the human kinome provides an extensive resource for promiscuity analysis and the exploration of inhibitor-based kinase relationships. Systematic organization of these relationships enabled the identification of small sets of kinases whose inhibitor binding profiles are representative of many others [1].
-
•
The data set is designed to complement kinase drug discovery projects in academia and the pharmaceutical industry by providing a wealth of inhibitor information for kinases of interest as well as template compounds for multi-kinase drug design.
-
•
Representative kinases can be used as primary targets for experimental evaluation of new inhibitors to estimate their potential for promiscuity across the human kinome. This substantially reduces initial experimental screening efforts. In addition, for individual kinase targets, the data set makes it possible to prioritize other kinases having very similar binding characteristics. These kinases can then be used as likely secondary targets to assess the potential selectivity of newly discovered kinase inhibitors.
1. Data description
The data set contains a total of 127,009 entries including 123,005 unique kinase-inhibitor interactions, 36,628 unique multi-kinase inhibitors, and 420 unique kinase targets. For each interaction entry, the following information is provided: Compound identifier (CPD_ID), “NonstereoAromatic” SMILES representation [3] of the inhibitor, standard potency measurement type, logarithmic potency value, full kinase name, standard abbreviation of the name, UniProt ID [11], kinase family of the target, kinase group [12], original source of the inhibitor, and a pan assay interference compound (PAINS) substructure alert, if it was detected using a PAINS filter [13]. The negative decadic logarithm of original potency measurements (IC50, Ki, or Kd), was calculated to yield the “pPot” value. Only inhibitors with pPot ≥ 6.0 were retained.
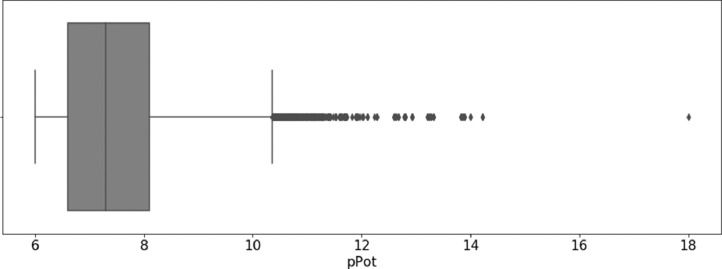
Fig. 1 shows the compound potency distribution within the data set. With a median value ˃ 7.0 more than half of the inhibitors are active in the nanomolar range.
Fig. 1.
Distribution of logarithmic potency values of data set compounds. The distribution is reported in a box plot. The vertical black line in the box indicates the median value.
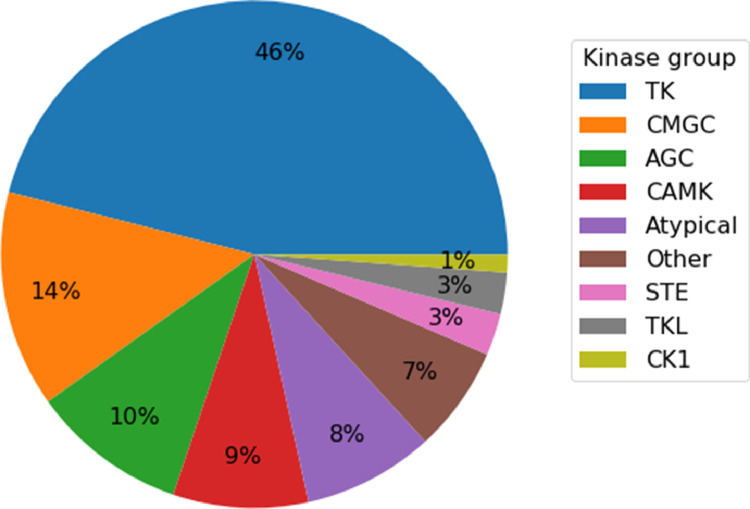
Fig. 2 shows the distribution of kinase-inhibitor interactions over kinase groups. With 46% of the interactions, tyrosine kinases (group TK) dominate the distribution, followed by serine‑threonine kinases (CMGC) with 14%. The remaining groups cover 1% - 10% of the interactions.
Fig. 2.
Distribution of kinase-inhibitor interactions across kinase groups. Nine kinase groups representing the human kinome are listed and color-coded according to the pie chart.
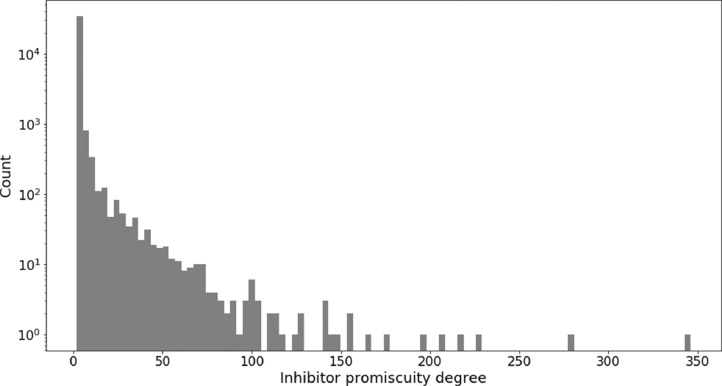
Fig. 3 shows the distribution of the promiscuity degree (number of kinase annotations) of the inhibitors. The data set contains 32 pan-kinome inhibitors with 100 to 346 kinase annotations. In addition, there are 453 inhibitors with 20 to 99 kinase annotations. However, most multi-kinase inhibitors have low to moderate promiscuity. Specifically, 98.7% (36,143) of the inhibitors comprising the data set are active against at most 10 kinases including 59.1% (21,650) with reported dual-kinase activity. The predominance of multi-kinase inhibitors with low promiscuity degrees mirrors the observed global distribution of promiscuous bioactive compounds [14].
Fig. 3.
Promiscuity degree of kinase inhibitors. The histogram reports the number of kinase inhibitors (y-axis, Count, on a logarithmic scale) with increasing promiscuity degree (x-axis, Inhibitor promiscuity degree). The promiscuity degree corresponds to the number of kinases an inhibitor is active against.
2. Experimental design, materials and methods
The source of multi-kinase inhibitor data set was an in-house compiled database of human kinase inhibitors assembled from public repositories comprising 112,624 unique inhibitors with activity against 426 kinases [1]. As potency measurements, IC50, Ki, or Kd values were mostly used. For generating the data set reported herein, we only selected inhibitors with multi-kinase activity applying a general potency threshold of 1 µM (pPot = 6.0), resulting in 36,628 inhibitors with activity against 420 kinases, forming a total of 123,005 unique inhibitor-kinase interactions. The data set is made available in standard .csv format.
All data selection and preparation steps were carried out using a customized Python script covering the following six steps:
(1) Load all required libraries, (2) define functions, (3) load source data, (4) specify filtering variables, (5) Identify multi-target inhibitors and process activity data, (6) generate plots (Fig. 1-3).
The Python code is also made freely available together with the data set.
Ethics statement
This is a secondary data set and thus did not involve any human or animal testing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgment
The authors are grateful to Filip Miljković for assembling the in-house source database.
References
- 1.Laufkoetter O., Laufer S., Bajorath J. Identifying representative kinases for inhibitor evaluation via systematic analysis of compound-based target relationships. Eur. J. Med. Chem. 2020;204:112641. doi: 10.1016/j.ejmech.2020.112641. [DOI] [PubMed] [Google Scholar]
- 2.Miljković F., Bajorath J. Computational analysis of kinase inhibitors identifies promiscuity cliffs across the human kinome. ACS Omega. 2018;3:17295–17308. doi: 10.1021/acsomega.7b01960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. [Google Scholar]
- 4.Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibrián-Uhalte E., Davies M., Dedman N., Karlsson A., Magariños M.P., Overington J.P., Papadatos G., Smit I., Leach A.R. The ChEMBLDatabase in 2017. Nucleic Acids Res. 2017;45:D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., Wang J., Yu B., Zhang J., Bryant S.H. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skuta C., Popr M., Muller T., Jindrich J., Kahle M., Sedlak D., Svozil D., Bartunek P. Probes & drugs portal: an interactive, opendata resource for chemical biology. Nat. Meth. 2017;14:759–760. doi: 10.1038/nmeth.4365. [DOI] [PubMed] [Google Scholar]
- 7.Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang R., Fang X., Lu Y., Yang C.Y., Wang S. The PDBbind database: methodologies and updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt T., Samaras P., Frejno M., Gessulat S., Barnert M., Kienegger H., Krcmar H., Schlegl J., Ehrlich H.C., Aiche S., Kuster B., Wilhelm M. ProteomicsDB. Nucleic Acids Res. 2018;46:D1271–D1281. doi: 10.1093/nar/gkx1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang J., Tanoli Z.-U.-R., Ravikumar B., Alam Z., Rebane A., Vähä-Koskela M., Peddinti G., van Adrichem A.J., Wakkinen J., Jaiswal A., Karjalainen E., Gautam P., He L., Parri E., Khan S., Gupta A., Ali M., Yetukuri L., Gustavsson A.-.L., Seashore-Ludlow B., Hersey A., Leach A.R., Overington J.P., Repasky G., Wennerberg K., Aittokallio T. Drug target commons: a community effort to build a consensus knowledge base for drug-target interactions. Cell. Chem. Biol. 2018;25:224–229. doi: 10.1016/j.chembiol.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.UniProt Consortium, UniProt A hub for protein information. Nucleic Acids Res. 2002;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manning G., Whyte D.B., Martinez R., Hunter T., S The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 13.Baell J.B., A.Holloway G. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y., Bajorath J. Entering the ‘big data’ era in medicinal chemistry: molecular promiscuity analysis revisited. Future Sci. OA. 2017;3:FSO179. doi: 10.4155/fsoa-2017-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]