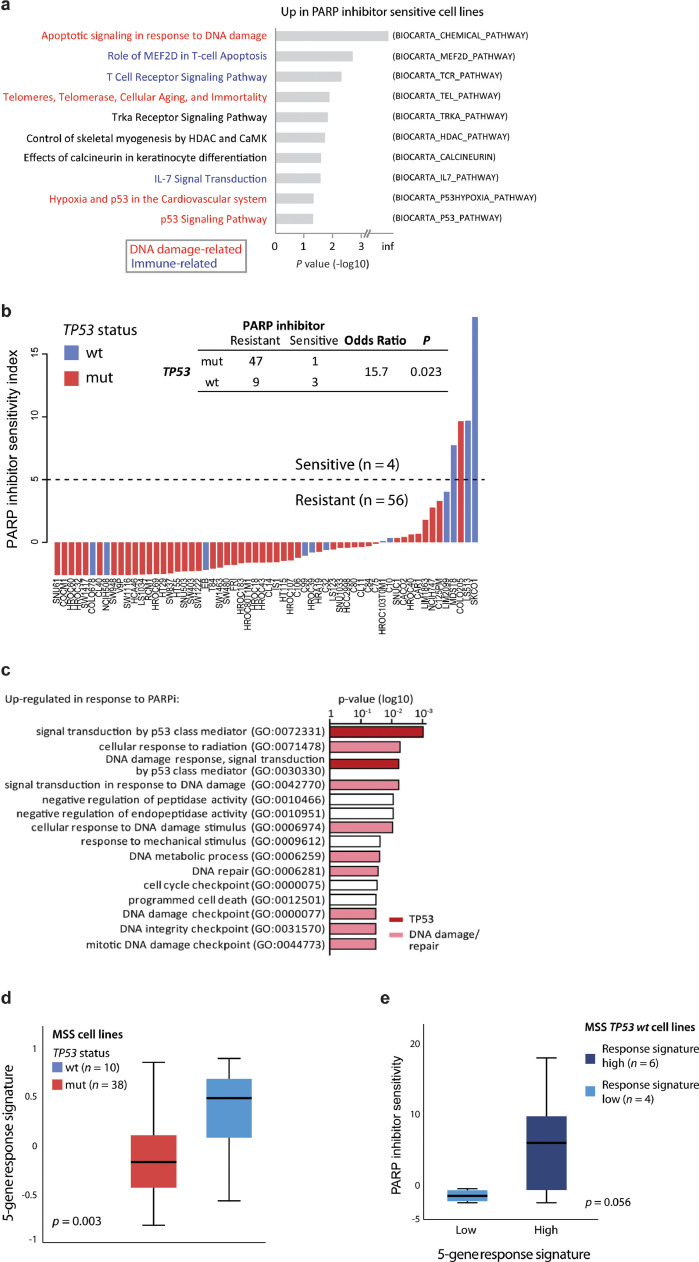
Fig. 4.
PARP inhibitor sensitivity is associated with TP53 activity. a Gene set enrichment analysis comparing PARP inhibitor resistant (n = 53) and sensitive (n = 4) colorectal cancer cell lines. Only microsatellite stable cell lines are included. Gene sets from BioCarta. The ten most upregulated gene sets in PARP inhibitor sensitive cell lines are shown, ranked according to p values (log10-scale). b Barplot shows summed standardized PARP inhibitor drug sensitivity scores per cell line, ranked according to drug sensitivity and coloured according to TP53 mutation status. Dashed horizontal line indicates value used for dichotomy in downstream analyses. Only MSS cell lines with known TP53 mutation status are included (n = 60). Similar plot including MSI cell lines is shown in Supplementary Fig. S3A. c Gene set enrichment analysis of genes associated with PARP inhibitor response. Analysis based on genes upregulated (paired limma-analysis) in PARP inhibitor sensitive cell lines (SKCO1 and LS513) after treatment with talazoparib compared to DMSO (control). d Five-gene response signature (GDF15, PLK2, MDM2, TP53INP1, RRM2B) according to TP53 mutation status in untreated MSS cell lines. e Comparison of PARP inhibitor sensitivity scores in MSS TP53 wild-type (wt) cell lines split according to the 5-gene response signature. Samples are dichotomized according to high or low gene response signature using a threshold of 0.3, reflecting the largest discontinuity in the distribution of response signature values. P value from Welch's t-test.