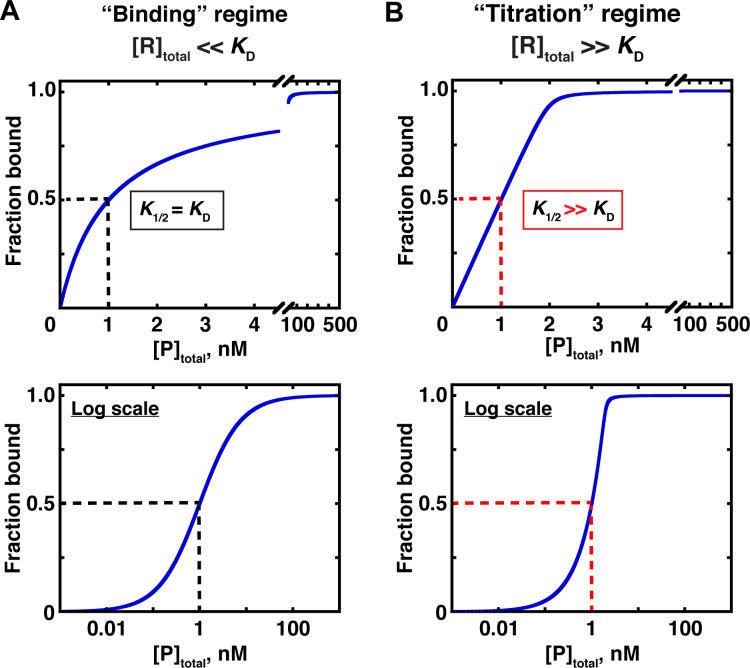
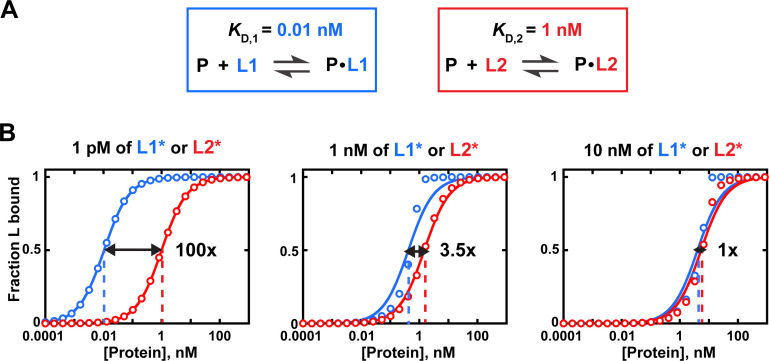
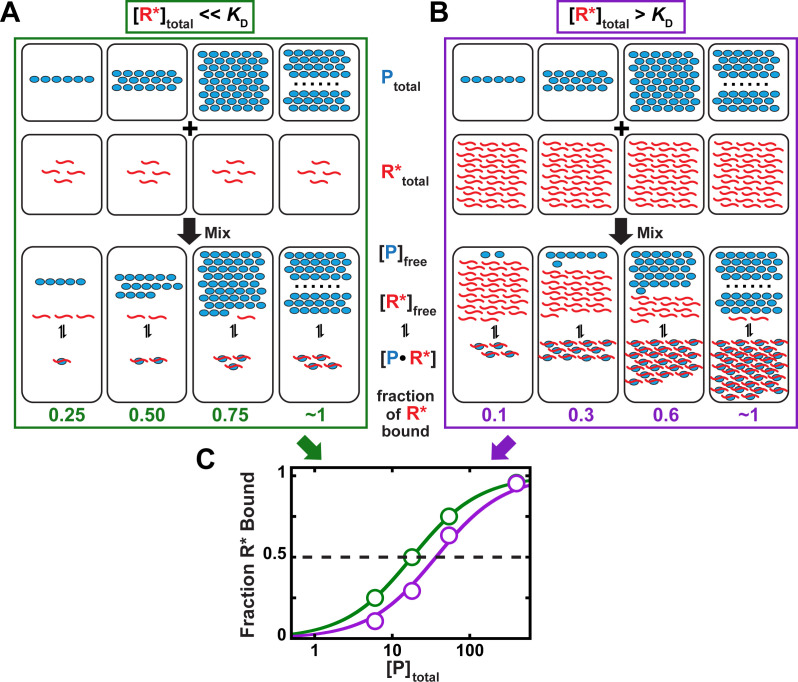
(
A) Labeled RNA concentration is much lower than
KD ([R*]
total <<
KD; binding regime). (
B) Labeled RNA concentration is greater than
KD ([R*]
total >
KD; intermediate regime). In parts (
A) and (
B), concentrations are indicated schematically by the number of RNA (R*, red), protein (P, light blue) molecules and RNA-protein complexes (P●R*) shown. In each case, protein concentration is varied (6, 18, 54, 400 arbitrary units), and
KD equals 18 (in the same units). The total RNA concentration is 4 (
A) and 36 (
B). (
C) Protein concentration dependence of binding in each of the above regimes. In the binding regime (green, [R*]
total <<
KD from part
A), the protein concentration at which half of the RNA is bound corresponds to the
KD. In contrast, in the intermediate regime (purple, [R*]
total >
KD from part
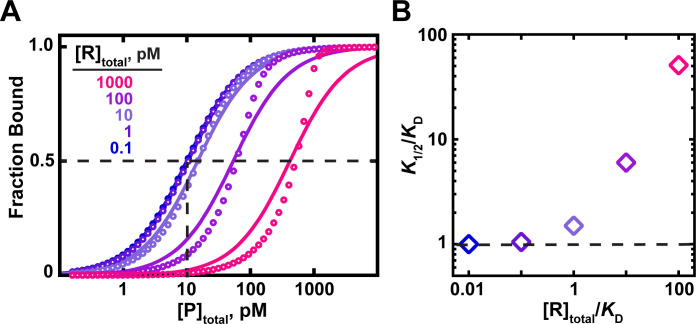
B), a greater protein concentration is required to achieve half-saturation (40 vs. 18 arbitrary units). The discrepancy would further increase with higher RNA concentrations, as shown in
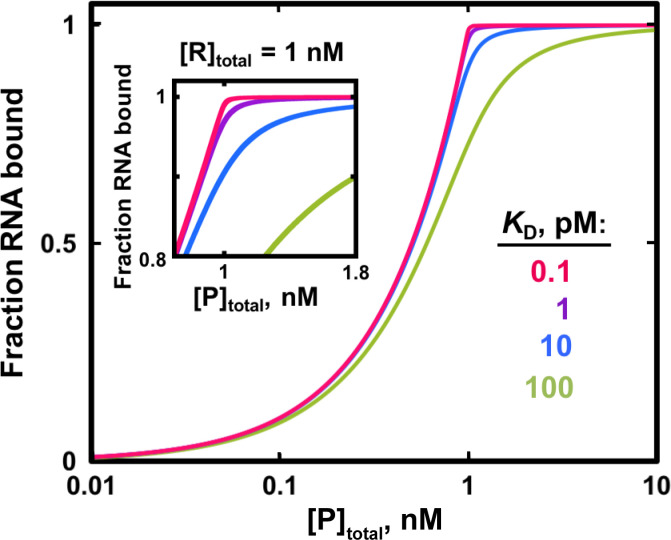
Figure 5—figure supplement 1. We can understand the origin of this discrepancy as follows. In part (
A), the RNA concentration (red) is below the
KD value and below the protein concentration (blue), such that the free concentration of the protein is essentially unchanged after RNA binding at both saturating (complete binding of RNA) and sub-saturating protein concentrations. Changing the RNA concentration in this regime would not change the fraction of RNA bound at a given total protein concentration, as long as the [R*]
total <<
KD condition remains met. On the contrary, in part (
B), the RNA concentration exceeds the dissociation constant (
KD) and is high enough that a large fraction of the total protein is bound by RNA. Thus, the
free protein concentration, which determines the extent of binding according to
Equation 4a, is depleted and can no longer be approximated by the
total protein concentration in
Equation 4b to obtain an accurate
KD value. On the molecular scale, the lowered free protein results in less binding. Consequently, for a given
KD, more protein is required to achieve half-saturation at higher RNA concentration than with a trace concentration of RNA. Intuitively, at a concentration of RNA that is greater than
KD there simply isn’t enough protein to occupy half the RNA when the total protein concentration is equal to
KD.