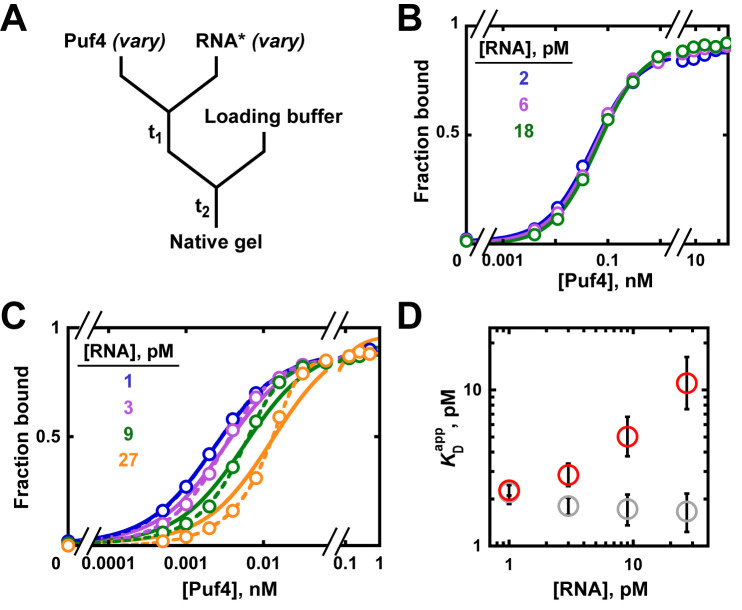
Figure 6. Varying the concentration of the 'trace' binding partner.
(A) Mixing scheme, as in Figure 4A but now with a series of labeled RNA concentrations. (B) Puf4 binding to different concentrations of 32P-labeled RNA at 25°C. For simplicity, only the lower limits of RNA concentration are indicated; the corresponding upper limits were 15–140 pM RNA (see Materials and methods and Appendix 2—note 4). Incubation time t1 was 0.5 hr, as established in Figure 4B. (C) Puf4 binding to different concentrations of 32P-labeled RNA at 0°C. Lower limits of labeled RNA concentration are indicated. Incubation time t1 was 40 hr. Note that these data are not fit well by Equation 4b, which assumes [R*]total << KD (solid lines). Quadratic fits, which do not assume negligible RNA concentration, are shown in dashed lines (Equation 5). (D) Effect of RNA concentration on apparent KD () at 0°C. Red symbols indicate values from a hyperbolic fit (Equation 4b and solid lines in C) and grey symbols indicate values from fits to the quadratic equation (Equation 5). The error bars denote 95% confidence intervals, as determined by fitting the data to the indicated equation in Prism 8.