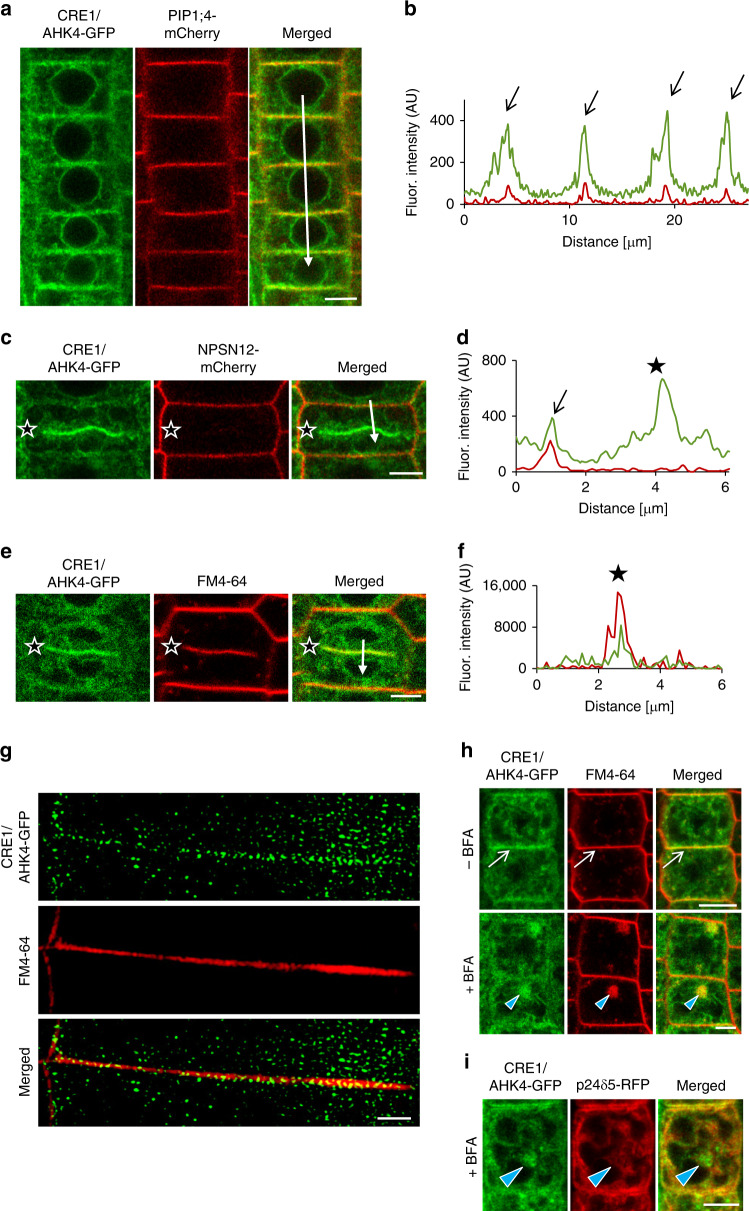
Fig. 4. CRE1/AHK4-GFP co-localization with the plasma membrane markers in Arabidopsis root cells.
a–f Co-localization of CRE1/AHK4-GFP with the plasma membrane markers PIP1;4-mCherry (a, b), NPSN12-mCherry (c, d) and FM4-64 (e, f) in the root meristematic epidermal cells. Profiles of fluorescence intensity of PM marker (red line) and CRE1/AHK4-GFP (green line) (b, d, f) were measured along the white lines (a, c, e) starting from the upper end (0 µm) towards the arrowhead. Peaks of PIP1;4-mCherry and NPSN12-mCherry fluorescence maxima correlate with the plasma membrane staining and overlap with CRE1/AHK4-GFP signal maxima (black arrows) (a–d). CRE1/AHK4-GFP signal detected at the cell plate of dividing cell (black stars on c, e) co-localizes with FM4-64 marker (e, f; black stars) but not with the plasma membrane marker NPSN12-mCherry (c, d; black stars). g Super-resolution imaging (SIM) of CRE1/AHK4-GFP subcellular co-localization with FM4-64 labelled PM. h, i Co-localization of CRE1/AHK4-GFP and FM4-64 (h), but not p24δ5-RFP (i) in endosomal compartments (blue arrowheads) formed in root meristematic epidermal cells treated with 50 µM in BFA for 1 h. Note CRE1/AHK4-GFP localization at the plasma membrane (white arrows) prior BFA treatment. Scale bars = 5 μm (a, c, e, h, i) and 2 μm (g).